Removal of synthetic dyes is one of the main challenges before releasing the wastes discharged by textile industries. Biodegradation of azo dyes by alkaliphilic bacterial consortium is one of the environmental-friendly methods used for the removal of dyes from textile effluents. Hence, this study presents isolation of a bacterial consortium from soil samples of saline environment and its use for the decolorization of azo dyes, Direct Blue 151 (DB 151) and Direct Red 31 (DR 31). The decolorization of azo dyes was studied at various concentrations (100–300mg/L). The bacterial consortium, when subjected to an application of 200mg/L of the dyes, decolorized DB 151 and DR 31 by 97.57% and 95.25% respectively, within 5 days. The growth of the bacterial consortium was optimized with pH, temperature, and carbon and nitrogen sources; and decolorization of azo dyes was analyzed. In this study, the decolorization efficiency of mixed dyes was improved with yeast extract and sucrose, which were used as nitrogen and carbon sources, respectively. Such an alkaliphilic bacterial consortium can be used in the removal of azo dyes from contaminated saline environment.
A number of colored effluents that contain dyes are released from textile, food, leather, dyestuff, and dyeing industries. The textile industry is one of the largest producers of effluents contaminated with dyes.1 The residual dyes released from these effluents introduce different organic pollutants in the natural water resources and land.2
Approximately 80,000tons of dyestuff and pigments are produced in India.1 It has been estimated that 10,000 different textile dyes are commercially available worldwide and the annual production is estimated to be 7×105 metric tons; 30% of these dyes are used in excess that is 1000tons per annum.3–5 During the dying process, about 2% of these dyes fail to bind to the substrate and are discharged in aqueous effluents.6 Azo dyes are the most widely used dyes in the industrial sector.7 They contain one or more azo groups (NN) that can resist the breakdown and accumulate in the environment at high levels with high degree of persistence.8,9
The wastewater from textile when directly released in the surface water without treatment can cause a rapid depletion of dissolved oxygen and lead to a great environmental damage.10 When dyes are available in the water system, the sunlight penetration into deeper layers is greatly reduced which disturbs photosynthetic activity resulting in deterioration of water quality, lowering the gas solubility, and finally causes acute toxic effects on aquatic flora and fauna. Most of the dyes that are released from wastewater, including their breakdown products, are toxic, carcinogenic, or mutagenic to humans and other life forms.11,12
Various physicochemical methods, such as adsorption on activated carbon, electrocoagulation, flocculation, froth flotation, ion exchange, membrane filtration, ozonation, and reverse osmosis, are used for the decolorization of dyes in wastewater. These methods are inefficient, expensive, have less applicability, and produce wastes in the form of sludge, which again needs to be disposed off.13
However, the microbial decolorization and degradation of azo dyes has gained considerable interest of researchers as it is inexpensive, eco-friendly, and produces less amount of sludge.14,15 It has been reported that many organisms are capable of reducing dyes, such as purely anaerobic (e.g., Bacteroides spp., Eubacterium spp., Clostridium spp.), facultatively anaerobic (e.g., Proteus vulgaris, Streptococcus faecalis), aerobic (e.g., Bacillus spp., Sphingomonas spp.), several yeasts, and even tissues from higher organisms.16–22
Effluents released from textiles industries are toxic, which contain a high degree of color (from residues of reactive dyes and chemicals) along with acidic and alkaline contaminants and high concentrations of organic materials.23 Extremophiles (alkaliphiles and halophiles) are metabolically diverse and can usually tolerate a greater amount of toxic metals and alkaline conditions in their environment.24 This study focuses on the decolorization of azo dyes by a moderately alkaliphilic bacterial consortium isolated from saline soil samples. The isolated bacterial consortium was used in the decolorization of azo dyes Direct Blue 151 (DB 151) and Direct Red 31 (DR 31) at different concentrations. The growth parameters for the consortium were optimized. The bacterial strains present in the consortium were identified by 16S rDNA sequencing.
Materials and methodsDyes and chemicalsThe textile dyes (azo dye compounds), namely DR 31 and DB 151, were purchased from the textile industry. Nutrient agar media and all other chemicals used in mineral salt medium (MSM) preparation were of analytical grade and purchased from Merck, India.
Bacterial consortium and culture conditionsThe bacterial consortium was isolated from soil samples of saline environment from three different regions of Chennai, namely Nagercoil, Tuticorin, and Pallavaram. The bacterial consortium was enriched in MSM amended with 100mg/L of DB 151 and DR 31. The composition of the MSM (pH 9) used for enrichment and decolorization was as follows: Na2HPO4: 12.8g/L; KH2PO4: 3g/L; NH4Cl: 1g/L; NaCl: 0.5g/L; 0.05M MgSO4: 10mL/L; 0.01M CaCl2: 10mL/L; and 20% glucose: 30mL/L.9 The medium was autoclaved, cooled, and then amended with 100mg/L of filter sterilized DB 151 and DR 31 in a 250mL Erlenmeyer flask. An amount of 10g of soil sample was aseptically inoculated into the medium. Individual bacterial isolates were obtained from the enriched culture by plating on nutrient agar medium containing 100mg/L of DB 151 and DR 31. The selected isolates were then purified by streaking on nutrient agar added with 100mg/L of the dyes. The single colony pure cultures were stored in 15% glycerol at 20°C.
Analytical techniquesAll decolorization experiments were carried out multiple times. MSM added with azo dyes was used as a control to determine abiotic color loss during the experiment. A volume of 1mL of precultured bacterial consortium was added to 50mL of MSM added with different concentrations (100, 150, 200, 250, and 300mg/L) of DB 151 and DR 31. The biodecolorization of DB 151 and DR 31 by bacterial consortium was observed for 5 days25. In order to monitor the decolorization process, the samples were withdrawn periodically, centrifuged at 10,000rpm for 15min, and filtered through syringe filter (PVDF, Millipore, Inc.); and decolorization was measured using UV/Vis spectra (Hitachi) at the corresponding λmax of the dye and was compared with the uninoculated control. The total protein content was also estimated at every 24h. The color removal efficiency of the bacterial consortium was determined as follows26:
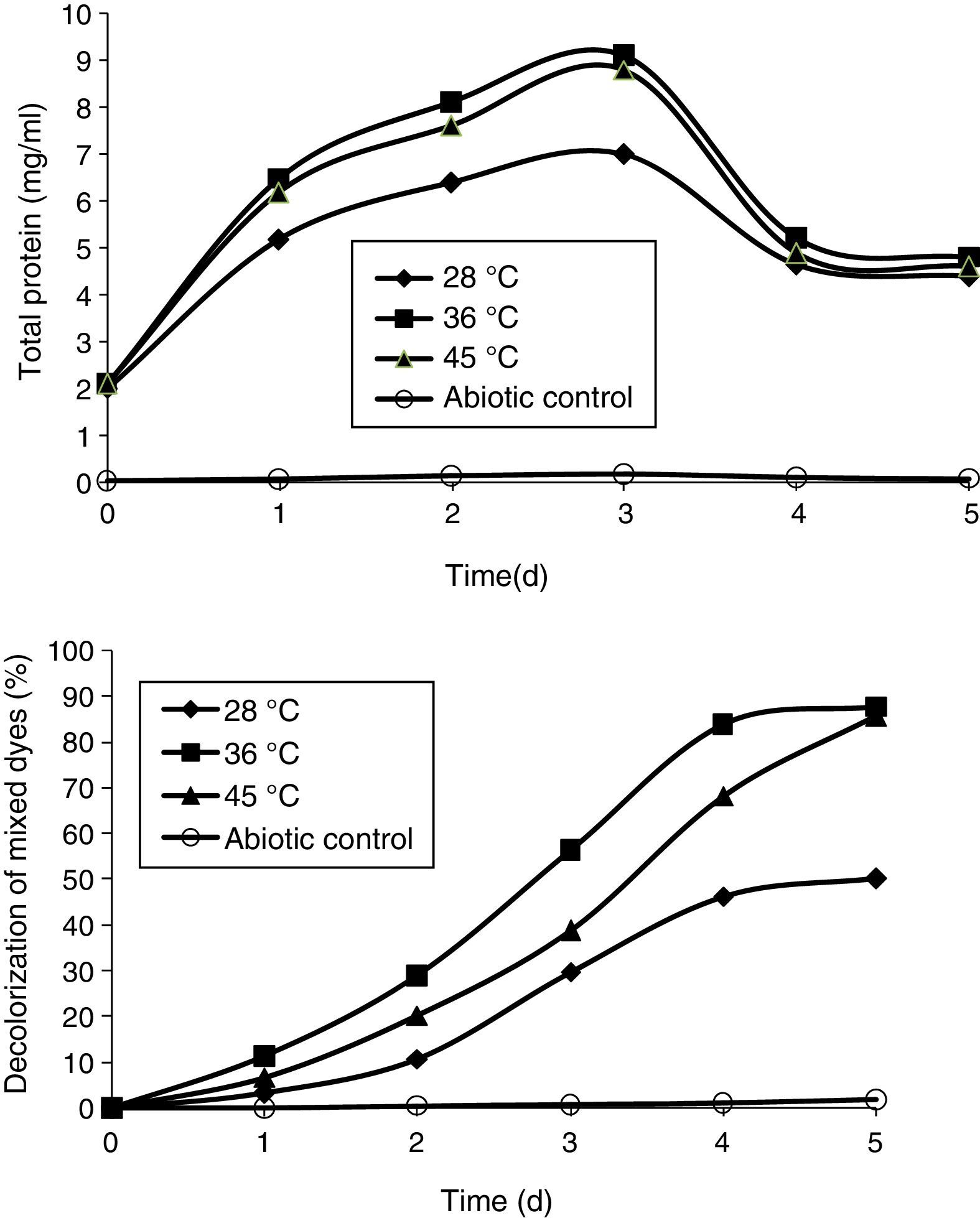
Effect of pH and temperature on the decolorization of mixed dyesIn order to study the effect of pH and temperature, the sterilized MSM was amended with 200mg/L of each of the DB 151 and DR 31 dyes. The medium was maintained at different pH: 8, 8.5, 9, 9.5, and 10. A volume of 1mL of overnight culture was inoculated in the flasks and incubated in a shaker at 36°C. The effect of temperature was studied by inoculating overnight culture and incubating in a shaker at 28°C, 36°C, and 45°C. The medium was maintained at pH 9.5. The measurement of decolorization of the total dye concentration was performed at an interval of 24h for 5 days.
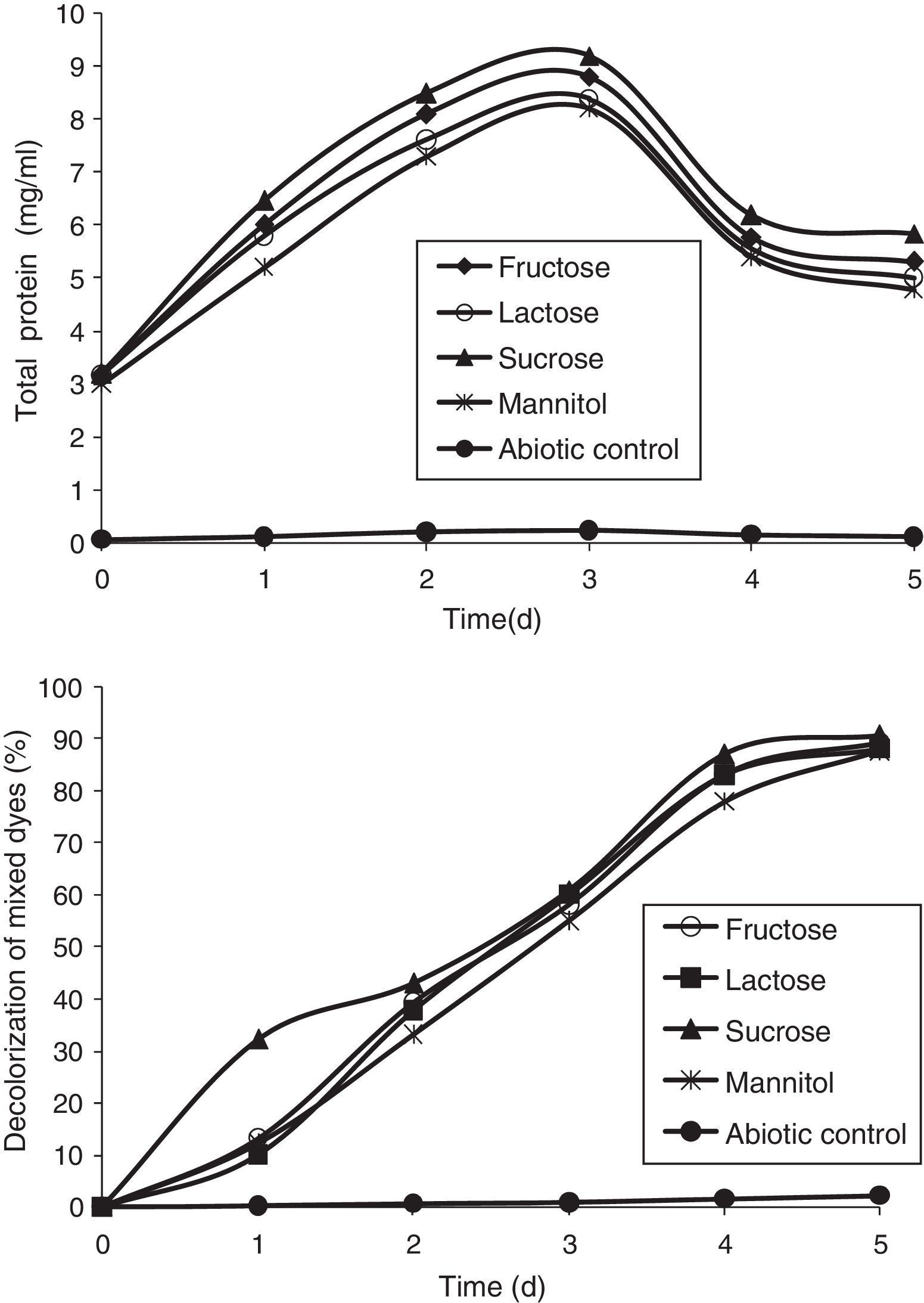
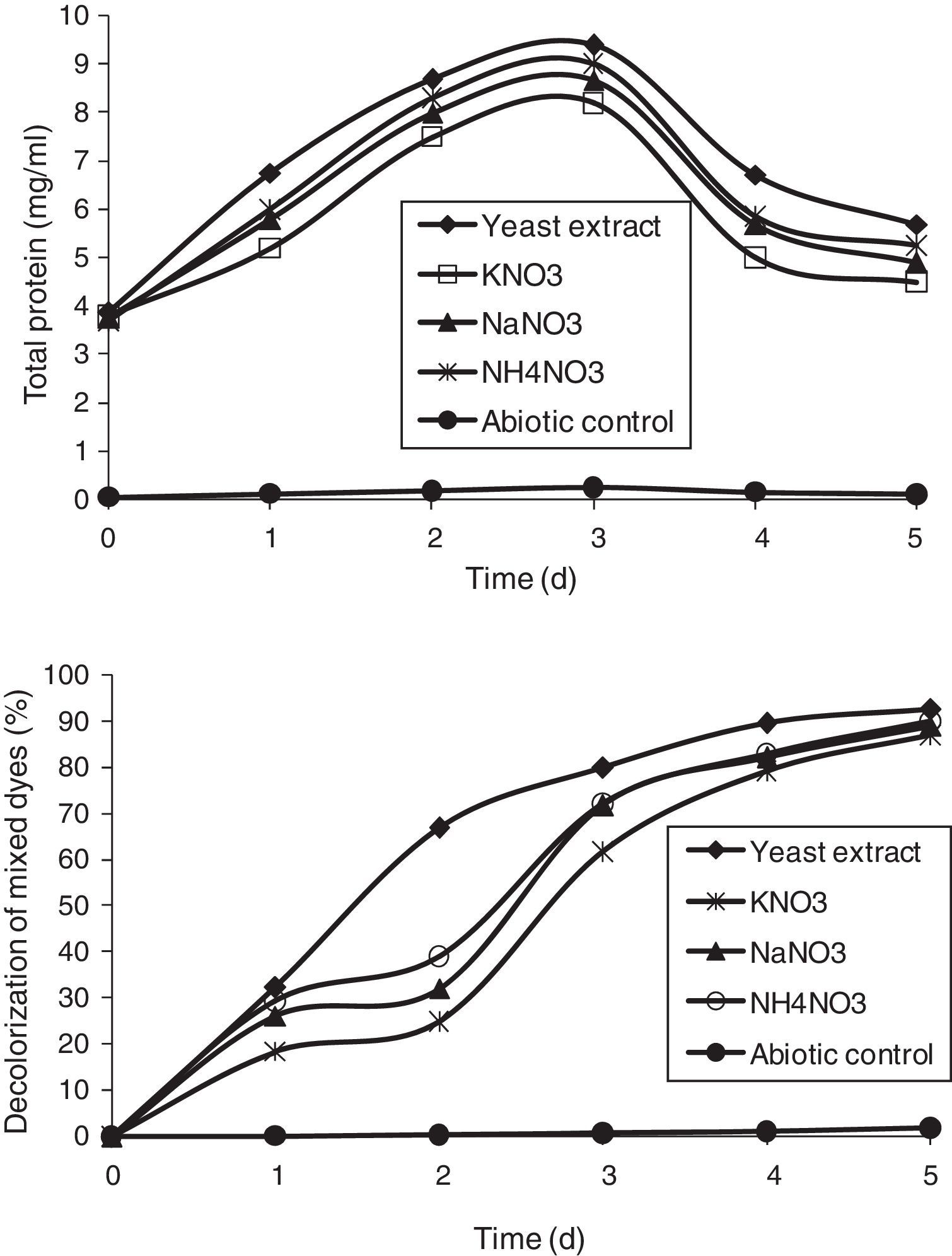
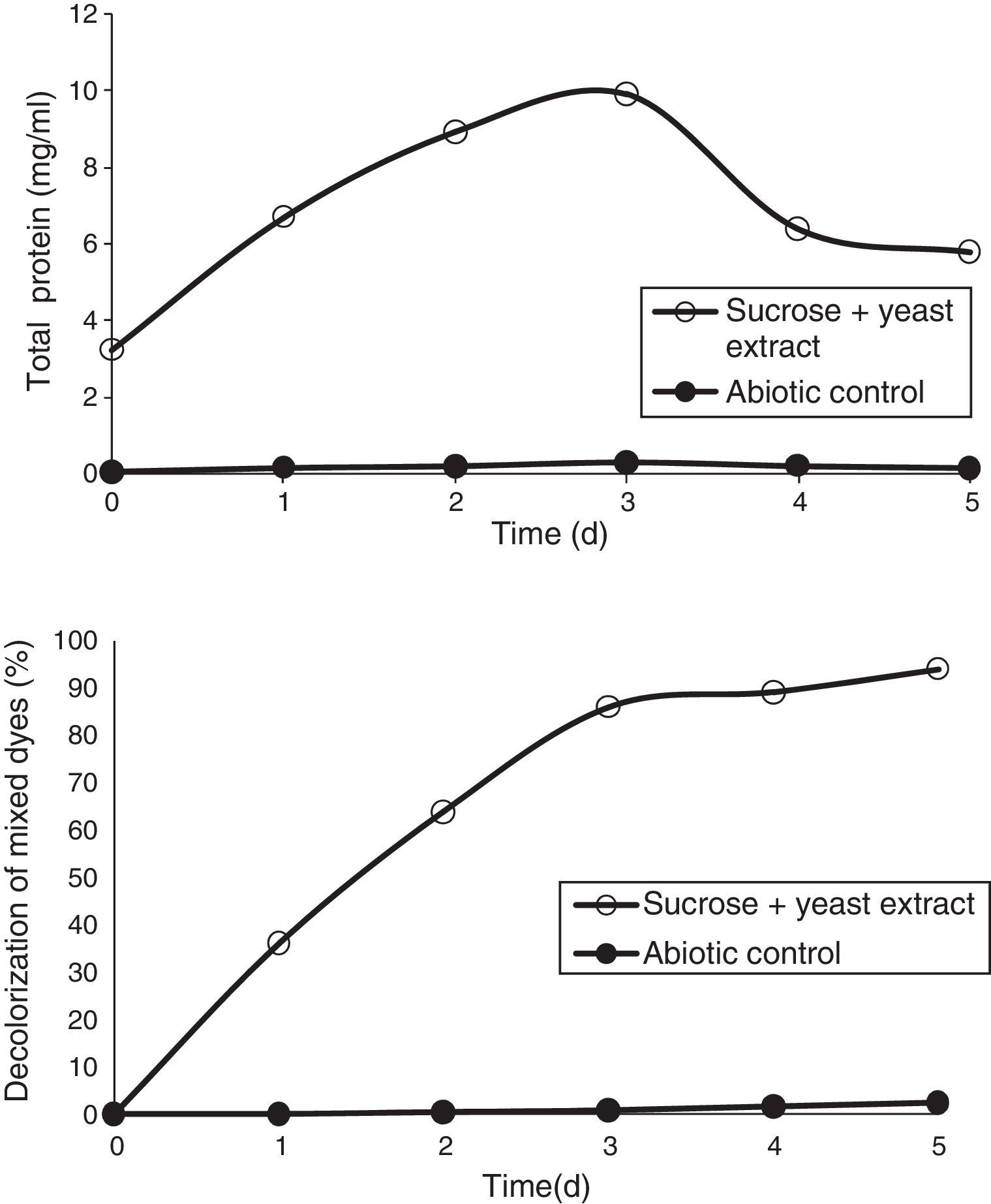
Effect of carbon and nitrogen sources on the decolorization of mixed dyesThe effect of carbon sources was studied using various compounds, such as fructose, lactose, sucrose, and mannitol, at a concentration of 1% and they were added individually as a supplement to MSM for the decolorization of mixed dyes. A volume of 1mL of the overnight culture was inoculated in the flasks and incubated in a shaker at 36°C. Nitrogen sources, such as yeast extract, KNO3, NaNO3, and NH4NO3 were added to MSM at a concentration of 0.5%; and 1mL of overnight culture was incubated at 36°C. In order to study the effect of efficient carbon and nitrogen sources, the optimum carbon and nitrogen sources, i.e., sucrose and yeast extract, were added to MSM at a concentration of 1% and 0.5%, respectively, and the decolorization of the dyes was measured.
Identification of azo-dye degrading bacterial strainsThe individual bacterial strains were separated from the consortium, and were used for the degradation of the azo dyes. The bacterial strains present in the consortium were initially examined using conventional biochemical tests. The molecular identification of the bacterial strains was performed by 16S rDNA sequencing. The bacterial strains present in the consortium were isolated and grown separately. Initially, Gram staining and motility tests were performed and then the biochemical characterization was carried out for different parameters (catalase, oxidase, indole production, citrate utilization, methyl red, Voges Proskauer, triple sugar iron agar, and urease) using 24h old culture of individual bacterial strains. After 24h of incubation at 37°C, the color change observed was accounted for a positive/negative result. The genus level identification of the unknown bacterial strains was carried out using Bergey's Manual of Systematic Bacteriology (2005) to ascertain the existence of variable biochemical test results for each strain.
16S rDNA partial gene sequencingChromosomal DNA was isolated from pure strains of the consortium by the standard phenol/chloroform extraction method (Sambrook). The 1.5kb partial sequence of 16S rDNA gene was amplified by the chromosomal DNA using polymerase chain reaction (PCR) with universal Eubacteria-specific primers 16F27 (5′-CCA GAG TTT GAT CMT GGC TCA G-3′) and 16R1525XP (5′-TTCTGCAGT CTA GAA GGA GGT GWT CCA GCC-3′).27 The used PCR conditions were: initial denaturation at 94°C for 2min, followed by 35 cycles of denaturation at 95°C for 1min, annealing at 55°C for 1min, extension at 72°C for 1min, and a final extension at 72°C for 10min; and finally sequencing was performed on an ABI310-automated DNA sequencer using Big Dye terminator kit (Applied Biosystems 3730xl DNA Analyzer). The amplified 16S rDNA gene (PCR products) from these isolates was directly sequenced after purification by precipitation with polyethylene glycol and NaCl. The primers used to obtain the complete sequence of 16S rDNA gene of the isolates were the same as used for the PCR amplification (16F27N and 16R1525XP).
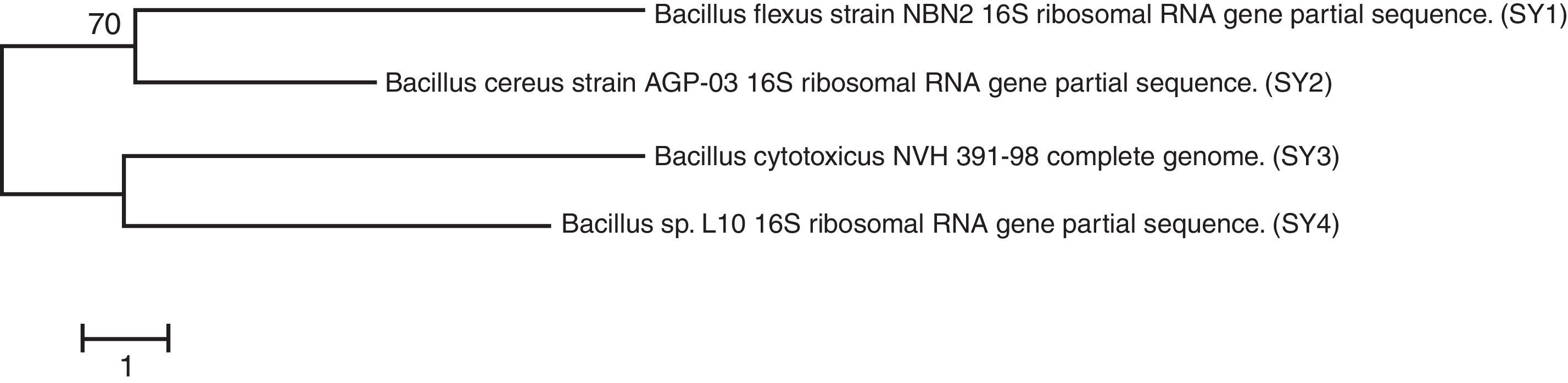
Sequence data analysis was performed using ChromasPro sequence analysis software. The phylogenetic tree was constructed by MEGA5.28 The evolutionary history was inferred using the neighbor-joining method. The optimal tree with the sum of a branch length of 0.18268090 is shown. The tree was drawn to scale with branch lengths (next to the branches) in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Kimura two-parameter method29 and were in the units of the number of base substitutions per site. The analysis involved three nucleotide sequences. The codon positions included were first, second, third noncoding sequences. All positions containing gaps and missing data were eliminated. There were a total of 1065 positions in the final dataset. The evolutionary analyses were conducted by MEGA5.30
Results and discussionAzo dyes are widely used in many industries. These azo dyes have been shown to be reductively cleaved by a wide range of microorganisms. Both aerobic and anaerobic bacteria from different environments possess the ability to reduce azo dyes into genotoxic compounds. This study aims to isolate alkaliphilic bacterial consortium that can be used for the removal of mixed dyes as textile wastes tend to have high pH.
Isolation and screening of azo-dye degrading bacterial consortiumThe initial enrichment of the bacterial consortium for the azo-dye degradation indicated four bacterial strains designated as SY1, SY2, SY3, and SY4 to be efficient. All of the four bacterial isolates were further grown on MSM-containing agar without addition of any carbon and nitrogen sources. They showed the ability to grow on the MSM agar after 48h of incubation at 37°C. The screening experiments for color removal were carried out under alkaline pH and aerobic conditions.
Sequence analyses of gene encoding for 16S rDNA from the bacterial isolatesThe phylogenetic tree (Fig. 1) constructed by MEGA530 displayed a relation between all the isolated bacterial strains. The isolates were identified as Bacillus flexus strain NBN2 (SY1), Bacillus cereus strain AGP-03 (SY2), Bacillus cytotoxicus NVH 391-98 (SY3), and Bacillus sp. L10 (SY4).
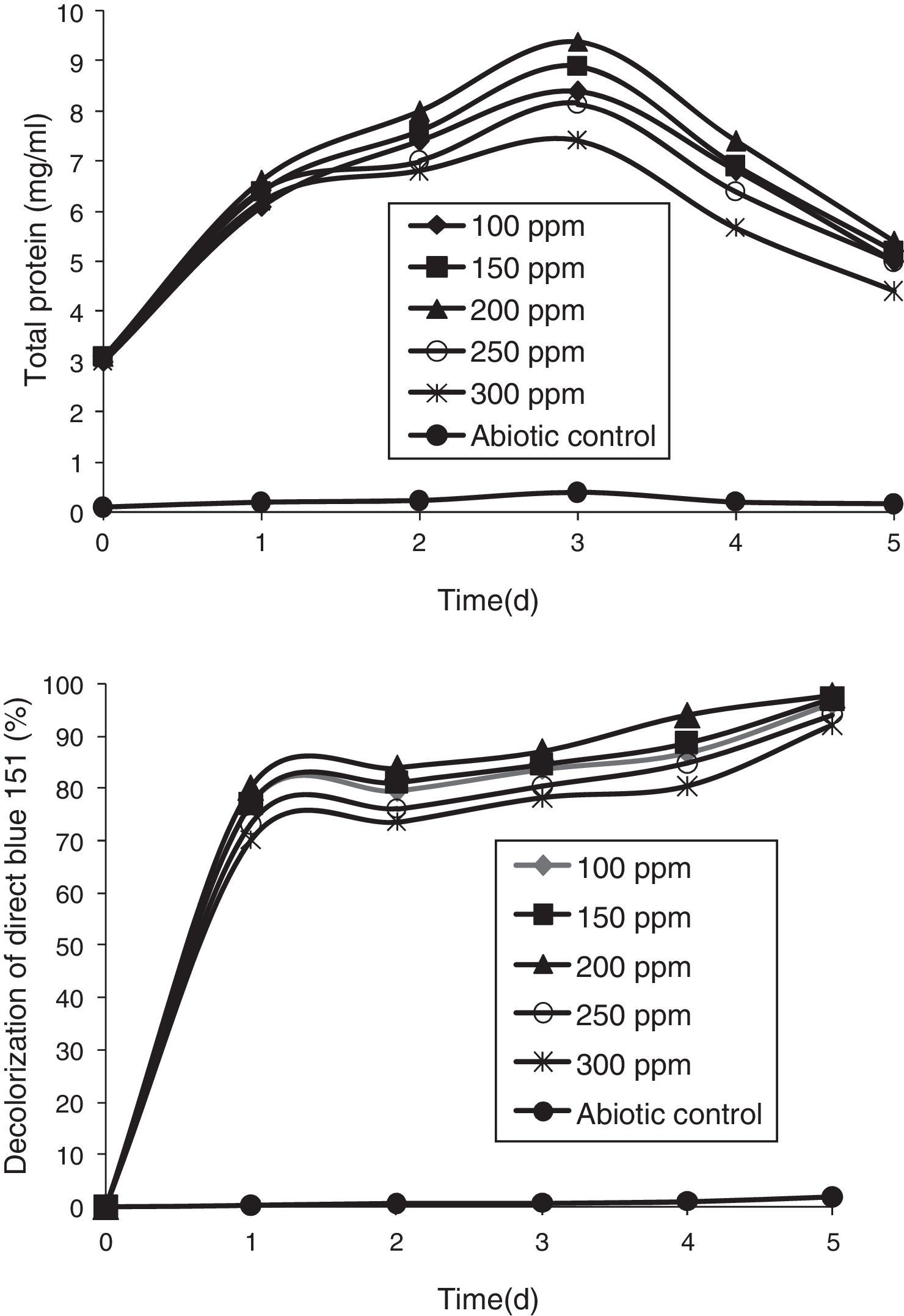
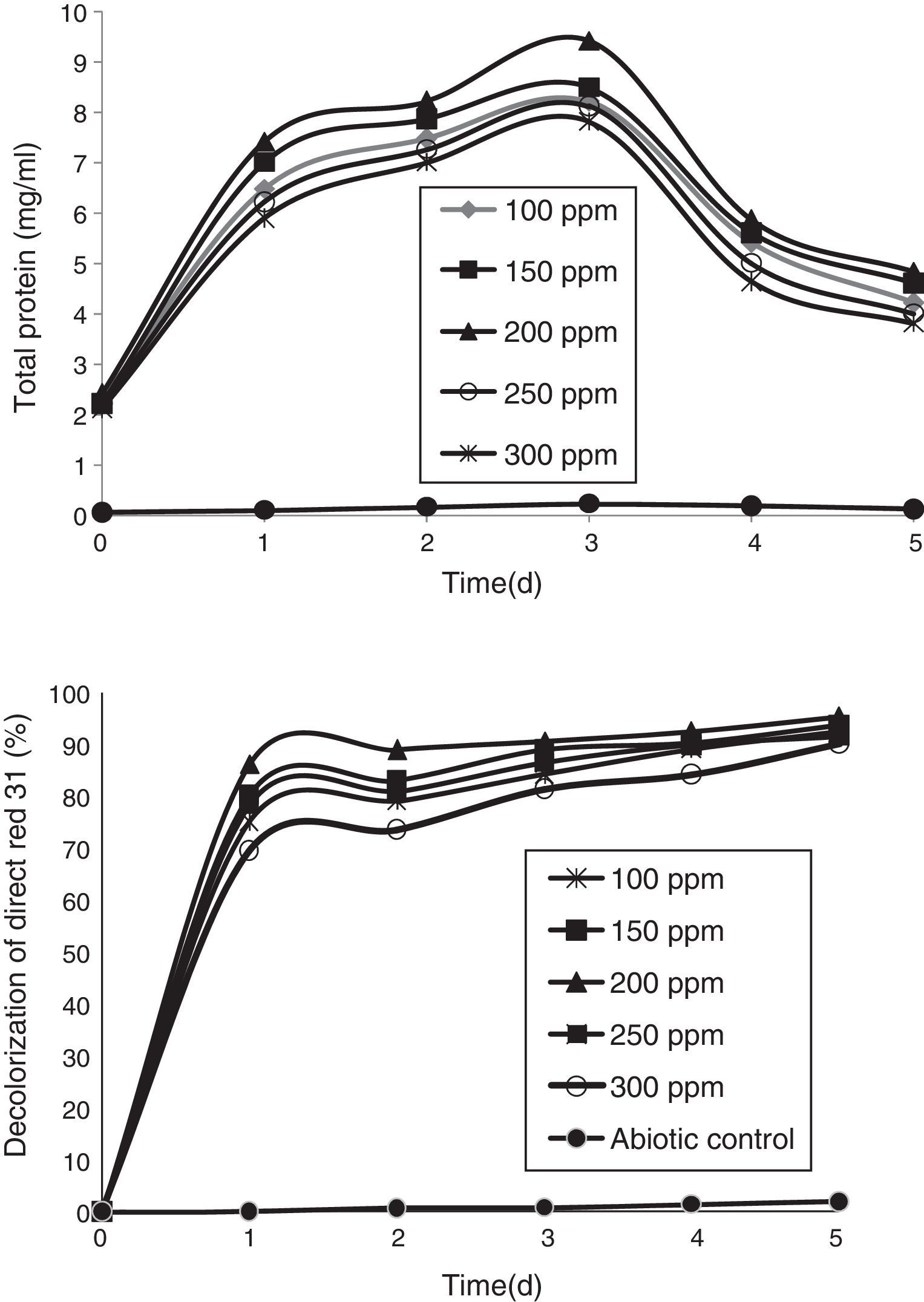
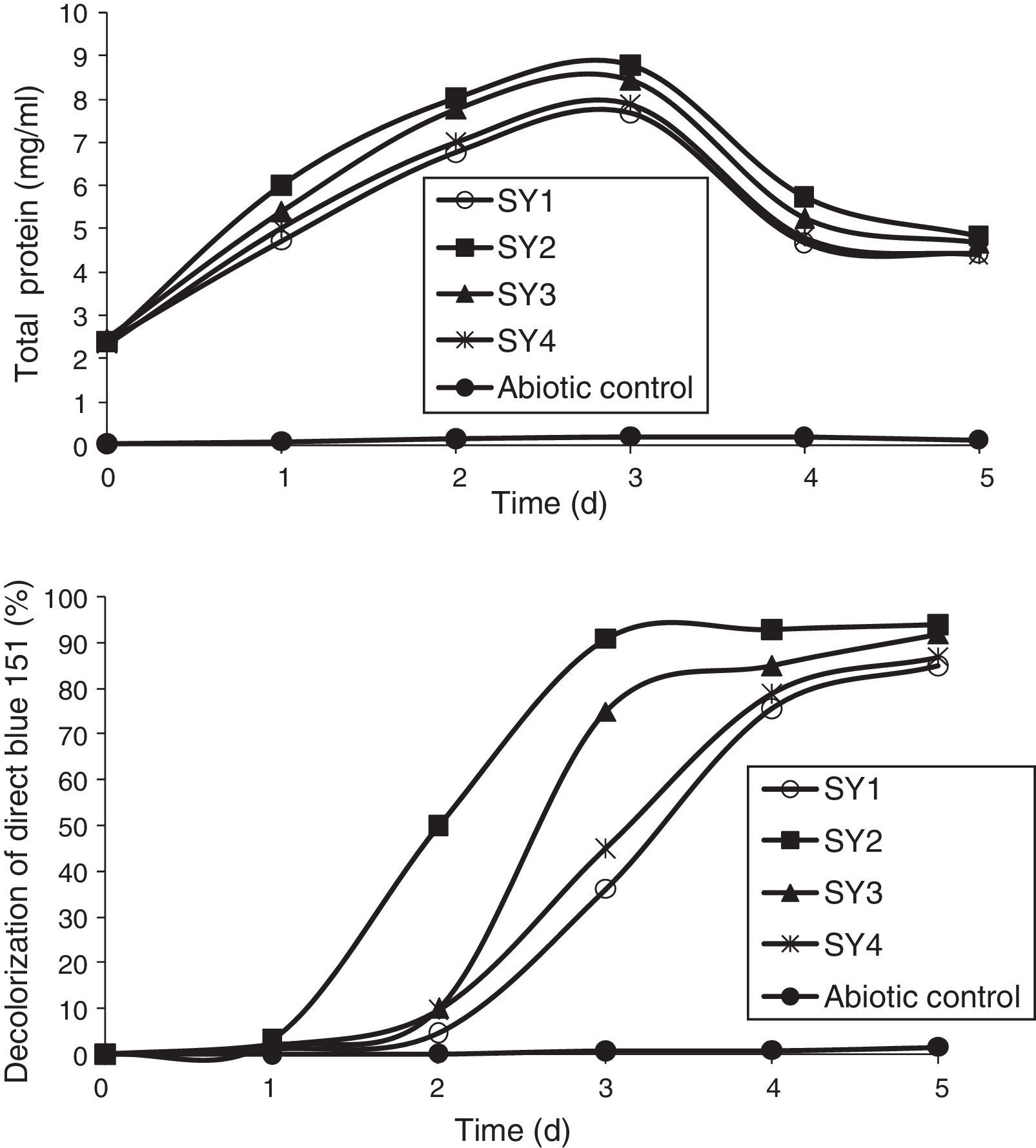
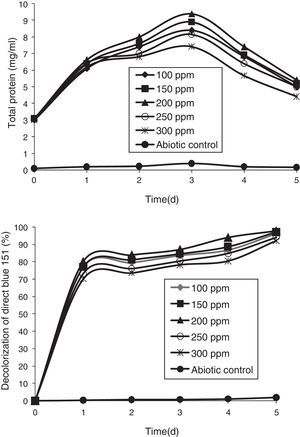
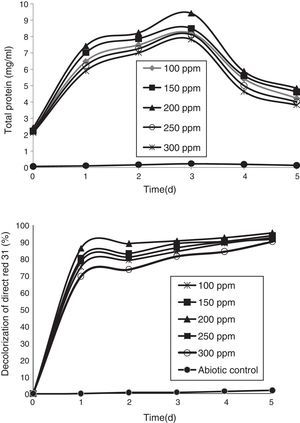
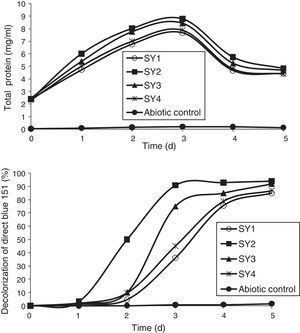
Azo-dye decolorization at various concentrationsThe bacterial consortium that contains the four strains enriched in the soil samples of saline environment was used for analyzing azo-dye decolorization. The ability of the bacterial consortium to decolorize the dyes DB 151 and DR 31, at various concentrations (100, 150, 200, 250, and 300mg/L) was investigated. The rate of decolorization increased with increase in initial dye concentration from 100 to 200mg/L, whereas decolorization decreased at 250mg/L which further continued to decrease at 300mg/L. The total protein content and decolorization at various concentrations of the dyes DB 151 and DR 31 are shown in Figs. 2 and 3.
Earlier studies on decolorization at various dye concentrations have been performed under non-alkaline conditions. These studies have shown the negative effect of increasing dye concentration from optimal level on dye decolorizing efficiency.31–34 This study was conducted under alkaline conditions. The decolorization of DB 151 and DR 31 increased from 100 to 150mg/L, i.e., 96% and 97% on the 5th day. The optimum concentration for efficient dye degradation was found to be 200mg/L for DB 151 and DR 31, where 97.57% and 95.25% of the dyes were degraded, respectively. The total protein content of the bacterial consortium increased, at 200mg/L of DB 151 and DR 31, from 6.62 to 9.4mg/mL and 7.41 to 9.41mg/mL from 1st to 3rd day, respectively. When the concentration of DB 151 was increased to 250mg/L, the decolorization of DB 151 and DR 31 was reduced to 93.99% and 91.56%, respectively. Decolorization at 300mg/L of DB 151 and DR 31 showed 91.89% and 90.08% reduction, respectively.
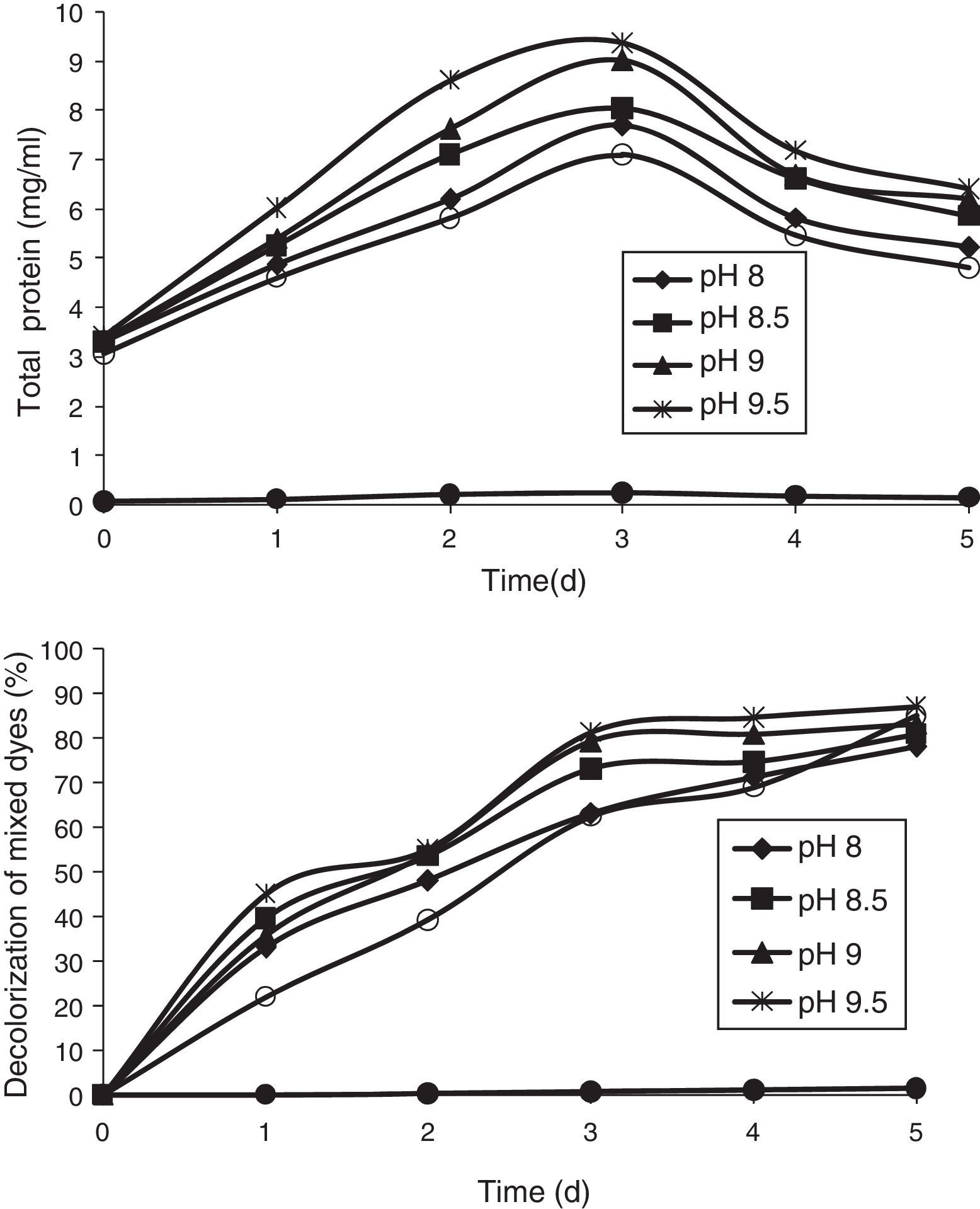
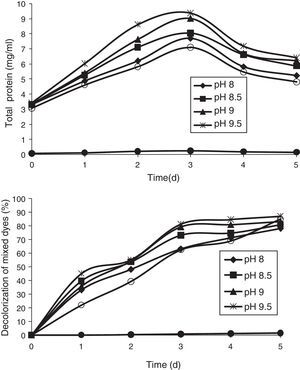
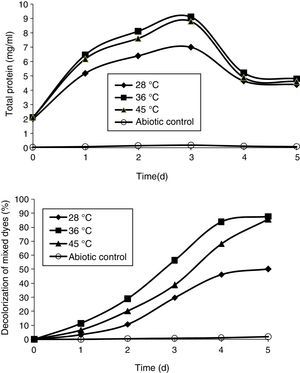
Effect of pHThe pH tolerance of decolorizing bacteria is quite important because reactive azo dyes are bound to cotton fibers by addition or substitution mechanisms under alkaline conditions and high temperatures.35 The effect of pH was studied at different pHs (8, 8.5, 9, 9.5, and 10). All the pHs allowed growth of the bacterial consortium. The maximum decolorization was observed at pH 9.5, which was 87% by the end of the 5th day. This was followed by pH 10, 9, 8.5, and 8, which showed 85%, 83.2%, 80.86%, and 78% of decolorization, respectively, by the end of the 5th day (Fig. 4).
Effect of temperatureThe effect of temperature was analyzed at 28°C, 36°C, and 45°C. The temperature 36°C enhanced the growth of the bacterial consortium and showed maximum decolorization of mixed dyes that was 87.49% by the end of the 5th day. This was followed by 45°C which showed decolorization up to 85.39%. The temperature 28°C showed the least decolorization of the mixed dyes that was 50% on the 5th day (Fig. 5). These results concord with an earlier study on the effect of various temperatures (25–40°C) on the decolorization of Fast Red by Bacillus subtilis, wherein the maximum decolorization was observed at 30–35°C.36
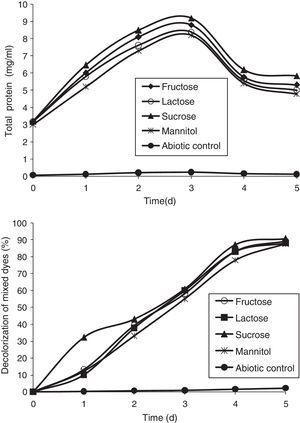
Effect of carbon sourcesTo examine the influence of carbon sources on the decolorization of mixed dyes (200mg/L), carbon sources such as lactose, sucrose, mannitol, and fructose, were supplemented in the media. It was found that sucrose could enhance the growth of the bacterial consortium more effectively than other carbon sources (Fig. 6). The decolorization of mixed dyes reached a maximum of 90.62% with sucrose as a carbon source followed by fructose, lactose, and mannitol which showed 89.08%, 88.11%, and 87.69% of decolorization, respectively. A similar observation was made by Ponraj et al.37 who reported the range of activity on the decolorization of Orange 3R with 1% sucrose as a carbon source by Bacillus sp., Klebsiella sp., Salmonella sp., and Pseudomonas sp., showing decolorization of 87.80%, 72.36%, 86.18%, and 80.49%, respectively, with the bacterium Bacillus sp. as the most effective decolorizer with sucrose as a carbon source.
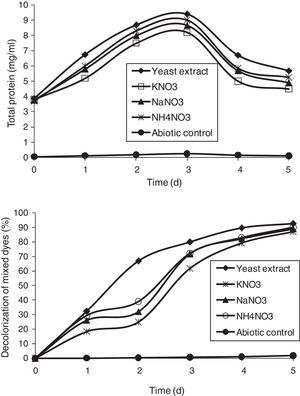
Effect of nitrogen sourcesOrganic nitrogen sources are considered essential media supplements for the regeneration of NADH that acts as an electron donor for the reduction of azo dyes by microorganisms.38 Nitrogen sources used in this study were 0.5% either of yeast extract, potassium nitrate (KNO3), sodium nitrate (NaNO3), and ammonium nitrate (NH4NO3), supplemented in MSM along with DB 151 and DR 31 at 200mg/L (Fig. 7). Yeast extract showed a maximum decolorization of 92.73% by the end of the 5th day, which was followed by NaNO3, NH4NO3, and KNO3. A report on the effect of various nitrogen sources, such as yeast extract, meat extract, peptone, and urea (0.5%) concurs with our study, wherein the used bacterial strains utilized yeast extract most effectively.7
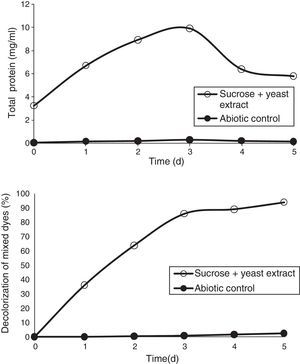
Effect of mixed carbon and nitrogen sources on the decolorization of mixed dyesTo increase the dye degradation efficiency, sucrose and yeast extract (1% and 0.5%) were added to the media. The addition of a combination of yeast extract and sucrose further enhanced the decolorization of mixed dyes by 94% at the end of the 5th day (Fig. 8).
Decolorization of mixed dyes by individual isolatesIndividual bacterial strains were analyzed for the decolorization of mixed dyes DB 151 and DR 31 at 200mg/L (Fig. 9). B. cereus showed maximum decolorization of 93.37% for mixed dyes under optimum conditions. This was followed by B. cytotoxicus, Bacillus sp. L10, and B. flexus showing 92.77%, 86.86%, and 85% decolorization, respectively. A study conducted by Ponraj et al.37 showed that Bacillus sp. has high decolorizing capacity for Orange 3R. They isolated Bacillus sp., Klebsiella sp., Salmonella sp., and Pseudomonas sp. from textile effluent and analyzed the decolorization of Orange 3R under optimum conditions and reported that Bacillus sp. and Pseudomonas sp. showed a similar level of decolorization (89%) followed by 80% and 76% by Salmonella sp. and Klebsiella sp., respectively.
ConclusionThis study reports that an enriched bacterial consortium can efficiently decolorize DB 151 and DR 31 up to 97.57% and 95.25%, respectively in 5 days. The bacterial consortium exhibited maximum decolorization ability of mixed dyes at pH 9.5. The physical parameters such as pH, temperature, and carbon and nitrogen sources play an important role in enhancing of the decolorization efficiency. The individual isolates were also able to degrade the mixed dyes. A strain of B. cereus showed maximum decolorization ability of up to 93.37% in 5 days. However, future work on the identification of genes present in the bacterial strains can be helpful in enhancing the decolorization of azo dyes.
Conflicts of interestThe authors declare no conflicts of interest.
Authors’ contributionsKVG conceived the study, participated in its design and drafted the manuscript; SY implemented the experimental work and helped in drafting the manuscript. All authors read and approved the final manuscript.