The present study attempts to examine the microbial profile and antibiotic susceptibility of diabetic foot infections in the intensive care unit of a tertiary referral centre for diabetic foot. As part of the study, we also attempted to find the prevalence of blaNDM-like gene among carbapenem-resistant gram negative infections.
MethodologyA prospective study of 261 patients with diabetic foot infections was performed during the period between January 2014 and June 2014.
ResultsA total of 289 isolates were obtained from 178 tissue samples from 261 patients, 156 (59.7%) males and 105 (40.2%) females, with a mean age of 58 years (−15 years), having diabetic foot infection. No growth was seen in thirty eight (17.6%) tissue samples. Out of the total samples, 44.3% were monomicrobial and 55.7% were polymicrobial. Gram negative pathogens were predominant (58.5%). Seven of the total isolates were fungal; 0.7% showed pure fungal growth and 1.7% were mixed, grown along with some bacteria. The most frequently isolated bacteria were Staphylococcus aureus (26.9%), followed by Pseudomonas aeruginosa (20.9%). Of the 58.5% gram negative pathogens, 16.5% were Enterobacteriaceae resistant to carbapenems. Among these isolates, 4 (25%) were positive for blaNDM-like gene. Among the rest, 18.6% were carbapenem-resistant Pseudomonas, among which 4 (36.3%) were blaNDM. Among the Staphylococci, 23.7% were methicillin-resistant Staphylococcus aureus.
ConclusionsOur results support the recent view that gram negative organisms, depending on the geographical location, may be predominant in DFIs. There is an increase in multidrug-resistant pathogens, especially carbapenem resistance and this is creeping rapidly. We need to be more judicious while using empiric antibiotics.
Foot ulcers and other foot problems are a major cause of morbidity and mortality in people with Diabetes mellitus.1 Diabetic foot infections (DFIs) are the leading cause of hospitalization for diabetic patients worldwide and in developing countries like India, it accounts for 20% of hospital admissions.2,3 DFI is a multifactorial process and three factors predispose to tissue damage, namely neuropathy, peripheral vascular disease, and susceptibility to infection whenever there is a direct injury to the foot at risk.4–6 DFIs are usually polymicrobial, caused by aerobic gram positive cocci like Staphylococcus aureus, gram negative bacilli (Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa), and anaerobes. Proper management of these infections needs appropriate antibiotic selection.7 Empirical treatment is based on the pathogens and the susceptibility pattern seen in the community where the hospital is located.
Beta-lactam antibiotics are the most commonly used antibiotics for bacterial infections.8 However, the accelerated emergence of antibiotic resistance to these groups of drugs among the prevalent pathogens is the most serious threat to the management of such infections, especially carbapenem resistance. These isolates are usually multidrug resistant, which further complicate the scenario.
There is a recent emergence of the NDM metallo-beta-lactamase (MBL) encoding genes among different enterobacterial species and also in non-fermenters like P. aeruginosa and Acinetobacter baumannii in various parts of world including India.9–11 In fact, India and Pakistan are the main reservoirs of blaNDM-like carrying Enterobacteriaceae.11 There is paucity of data on MBL-producing organisms carrying blaNDM-like gene from diabetic foot infections. Taking this into account, we studied the microbial profile and susceptibility pattern of diabetic foot infections in patients with Type 2 Diabetes mellitus to guide empiric therapy for diabetic foot infections in our hospital and the occurrence of blaNDM-like carbapenemase gene among carbapenem-resistant gram negative pathogens.
Materials and methodsStudy typeA prospective study was performed on 261 diabetic patients with foot ulcers over a period of six months from January 2014 to June 2014. The study was conducted at a tertiary care hospital in Mumbai, India.
Study populationAll patients with type 2 diabetes (irrespective of age and sex) who were hospitalized for surgical management of lower-extremity wounds from January 2014 till June 2014 were considered for the study. Their informed consent was obtained and demographic details, duration of lower-limb lesion, duration of diabetes, and type of empiric therapy were documented from their medical records. A deep tissue specimen was obtained from the wounds during surgery and sent for bacterial and fungal cultures.
Specimen collectionAfter surgical debridement of the slough and necrotic tissue over the wound in the operation theatre, the wound was washed thoroughly with normal saline; a deep tissue specimen of approximately 0.5×0.5cm was taken from the wound bed. The specimen was collected in a sterile container soaked with normal saline and was transported to our microbiology laboratory without delay for further processing.
Specimen processingPart of the sterile deep tissue specimen was crushed or ground with a sterile mortar and pestle in the biosafety cabinet. The crushed specimen was subjected to gram staining and was streaked on 5% sheep blood agar (SBA), MacConkey agar (MA), and Saboraud's Dextrose agar (SDA) for fungal culture. After inoculation, the SBA was kept in a candle jar and along with MA, it was kept in an incubator at 37°C. Bacterial isolates and yeast-like fungus identification and susceptibility test was performed using VITEK 2 Compact automated culture system (bioMeriux, France). Commercial I+ IE (imipenem+ imipenem/EDTA) disc from Himedia, Mumbai, India was used for EDTA disc synergy test. All carbapenem resistant Enterobacteriaceae were tested for metallobetalactamase production. Modified Hodge test (MHT) was performed for all Enterobacteriaceae isolates resistant to carbapenems by Vitek 2 according to CLSI guidelines and EDTA disc synergy test was done for the detection of metallo-β-lactamase production for all gram negative isolates. ESBL production was confirmed by ceftazidime and ceftazidime/clavulunic acid disc synergy test and detection of AmpC β-lactamases was performed as follows.
AmpC detection methodsThe isolates were screened for presumptive AmpC production by testing their susceptibility to cefoxitin (30μg) using Kirby Bauer disc diffusion method and interpreted according to the CLSI guidelines.12 All the isolates with an inhibition zone diameter of less than 18mm were labelled as screen positive.
A lawn culture of E. coli ATCC 25922 was prepared on MHA plate. A sterile disc of 6mm moistened with 20μL of sterile saline was kept and several colonies of test organism were inoculated on this disc. A cefoxitin disc was placed next to this disc (almost touching) on the inoculated plate. The plates were incubated overnight at 37°C. A flattening or indentation of the cefoxitin inhibition zone in the vicinity of the disc was considered a positive test.13
PCR technique for detection of blaNDMTotal DNAs of the different bacterial isolates were extracted by alkaline lysis and PCR to detect blaNDM-like gene was performed as described by Poirel et al.14
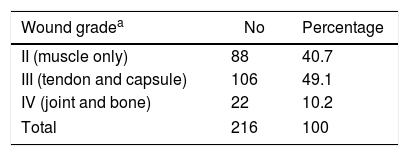
ResultsA total of 216 diabetic patients with foot ulcers admitted to our hospital were studied during the 6 months study period from January 2014 to June 2014. Out of the 261 patients, 156 [59.7%] were males and 105 [40.2%] females of mean age 58 years (±15 years) with a DFI. Majority of the DFIs were Grade III (49.1%) (Table 1).
Infections and depth of diabetic foot lesions.
| Wound gradea | No | Percentage |
|---|---|---|
| II (muscle only) | 88 | 40.7 |
| III (tendon and capsule) | 106 | 49.1 |
| IV (joint and bone) | 22 | 10.2 |
| Total | 216 | 100 |
A total of 289 isolates were obtained from 178 tissue samples. No growth was seen in thirty eight (17.6%) tissue samples. Distribution of the microorganisms in tissue samples is shown in Table 2. Monomicrobial infection was seen in 44.3% cases. Among the bacterial isolates, a total of 117 (41.5%) gram positive pathogens and 165 (58.5%) gram negative pathogens were isolated.
From the 165 gram negative isolates, 102 (61.8%) belonged to family Enterobacteriaceae. Among the Enterobacteriaceae isolated, Escherichia coli was found to be most common followed by Klebsiella pneumoniae (Table 3). Seven of the total isolates were fungal, of which 0.7% showed pure fungal growth and 1.7% were mixed, grown along with some bacteria. The fungal isolates corresponded to Candida albicans (2), Candida tropicalis (2), Candida spp. (1), Aspergillus fumigatus (1) and Rhodotorula sp.
Bacterial pathogens in diabetic foot infection.
| Name of the pathogen | Number of pathogens |
|---|---|
| Staphylococcus aureus | 76 (26.9%) |
| Pseudomonas aeruginosa | 59 (20.9%) |
| Enterococcus faecalis | 36 (12.7%) |
| Escherichia coli | 34 (12%) |
| Klebsiella pneumoniae | 27 (9.5%) |
| Citrobacter koseri | 10 (3.5%) |
| Proteus mirabilis | 9 (3.1%) |
| Enterobacter cloacae | 8 (2.8%) |
| Citrobacter freundii | 5 (1.7%) |
| Streptococcus pyogenes | 5 (1.7%) |
| Proteus vulgaris | 4 (1.4%) |
| Enterobacter aerogenes | 2 (0.7%) |
| Acinetobacter baumannii | 2 (0.7%) |
| Proteus penerri | 1 (0.3%) |
| Providencia stuartti | 1 (0.3%) |
| Serratia marcescens | 1 (0.3%) |
| Acinetobacter lwoffii | 1 (0.3%) |
| Stenotrophomonas maltophilia | 1 (0.3%) |
| Total | 282 |
Piperacillin-tazobactam and cefoperazone sulbactam were the most frequently used empiric antibiotics (68%) among DFI patients. The antibiotics used were of broad spectrum primarily because our hospital is a tertiary referral centre for diabetic foot infections and hence, we get many admissions from smaller setups and nursing homes.
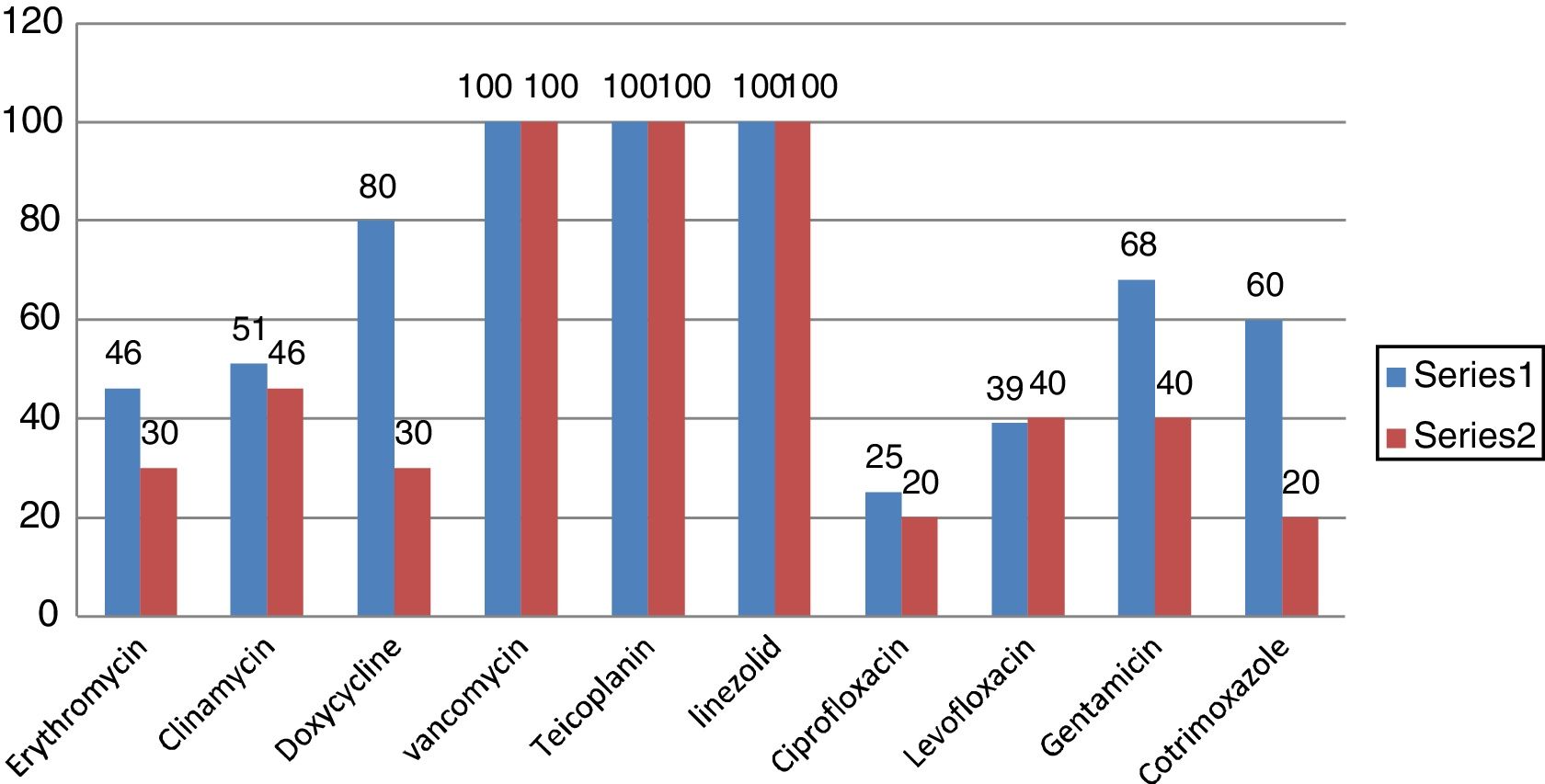
Out of 76 isolates, 18 (23.7%) were methicillin resistant S. aureus (MRSA). S aureus (MSSA +MRSA) strains were 100% sensitive to Vancomycin, Teicoplanin, and Linezolide (Fig. 1). The next common gram positive organism was Enterococcus faecalis and 11% (4 out of 36) were VRE (Vancomycin resistant Enterococcus) strains.
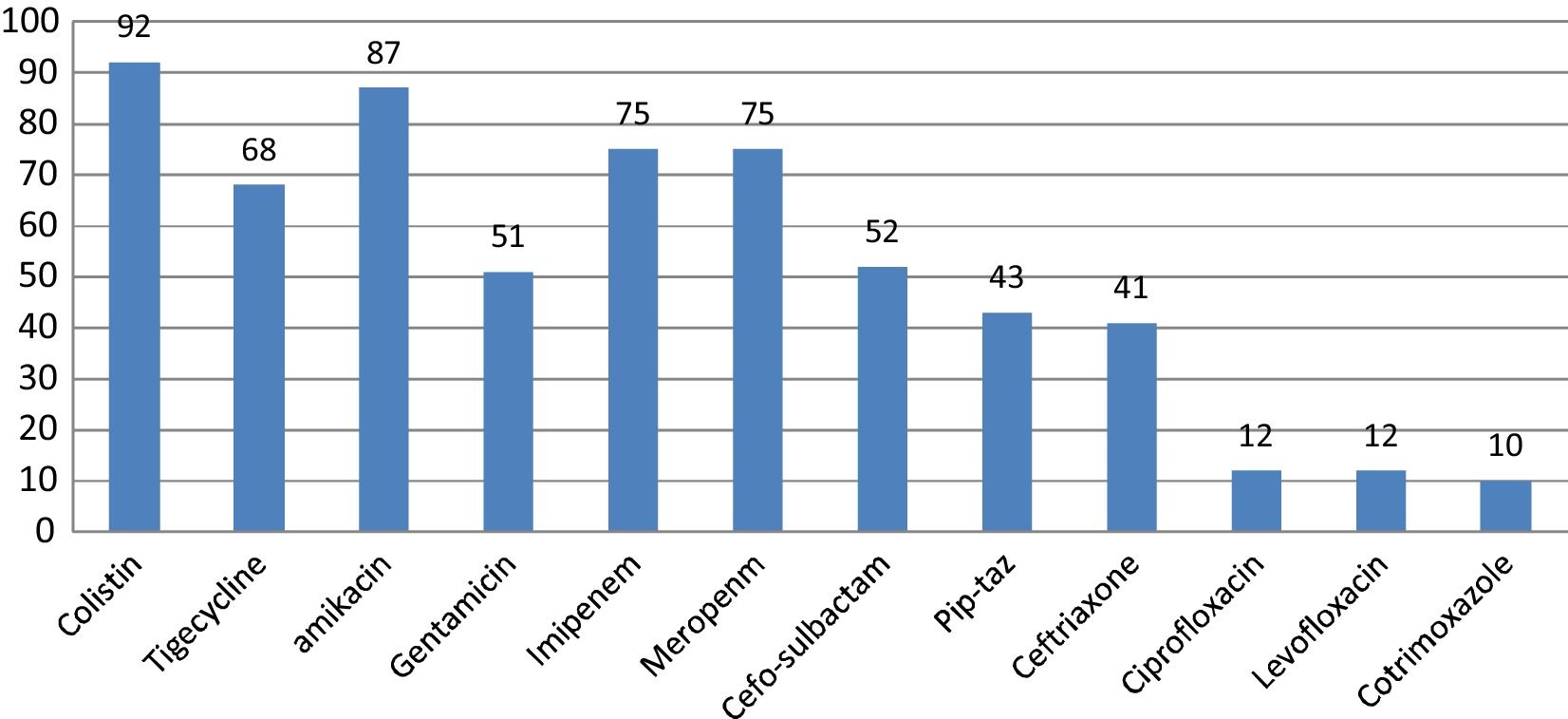
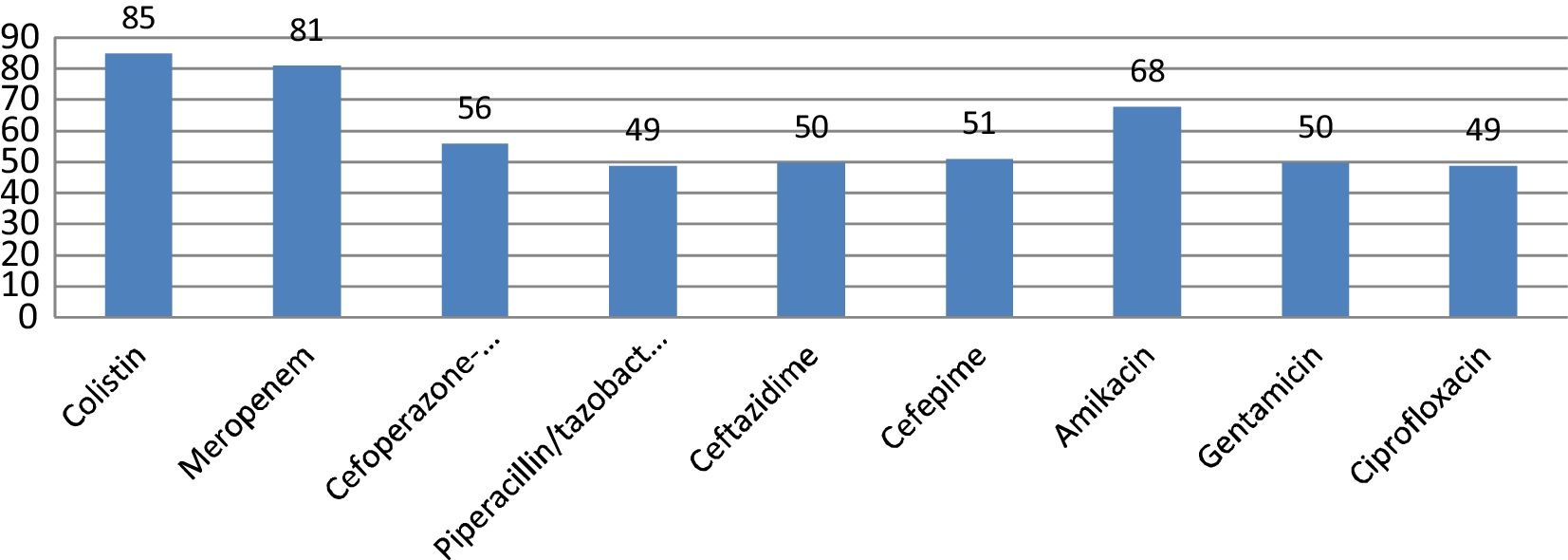
E. coli and K. pneumoniae were the most frequent Enterobacteriaceae isolated. Their susceptibility pattern is shown in Figs. 2 and 3. Colistin was the most effective agent against enterobacterial isolates, followed by aminoglycosides and beta-lactam/beta-lactamase inhibitor combinations. Two (7.4%) of the 27 Klebsiella isolates were resistant to colistin. E. coli isolates were 100% sensitive to colistin.
Among the Enterobacteriaceae, sixteen isolates (16.8%) were resistant to carbapenems (Table 4). Nine of the 16 (56.2%) carbapenem resistant strains were MHT positive and 7 of the 16 isolates were positive for MBL detection by EDTA disk synergy test (EDS). All 16 isolates were studied for molecular characterization for blaNDM-like gene. Four of the seven carbapenem-resistant K. pneumoniae and one of the 3 carbapenem-resistant E. coli isolates were positive for blaNDM-like gene. All the isolates positive for blaNDM like gene were positive by EDS but only one Klebsiella isolates harbouring blaNDM-like gene was positive by MHT (Table 4).
Phenotypic detection of resistance mechanisms in beta-lactam resistance Enterobacteriaceae (n: 95).
| ESBL | AmpC | CRE | MHT | EDS | |
|---|---|---|---|---|---|
| Klebsiella pneumoniae (n=27) | 9 (33.3%) | – | 7 (25.9%) | 5 (1-NDM +ve) | 4 (NDM +ve) |
| Escherichia coli (n=34) | 21 (61.7%) | 2 (5.8%) | 3 (8.8%) | 2 | 1 (NDM +ve) |
| Enterobacter cloacae (n=8) | 4 (50%) | – | 2 (25%) | – | – |
| Enterobacter aerogenes (n=2) | 1 (50%) | – | 1 (50%) | 1 | 0 |
| Citrobacter koseri (n=10) | 3 (30%) | – | – | – | – |
| Citrobacter freundii (n=5) | 2 (40%) | – | – | – | – |
| Proteus mirabilis (n=9) | 1 (11%) | - | 3 (33%) | 1 | 2 |
| 95 | 41 (43.1%) | 2 (2.1%) | 16 (16.8%) | 9 | 7 |
+ve means positive.
Among the gram negative isolates, 63 were non-fermenters being P. aeruginosa the most frequent (n=59[93.6%]). Eleven (18.6%) P. aeruginosa isolates were carbapenem resistant strains. Four (36.3%) of the 11 carbapenem resistant isolates were positive for blaNDM-like gene, all 4 were positive by EDS. Fifteen percent (9/59) of the P. aeruginosa, were resistant to colistin. There were isolated two A. baumannii cases and both were carbapenem resistant, blaNDM positive, and susceptible only to colistin and tigecycline (Fig. 4).
Outcome of patients with DFIAmputation of the affected part was done in 120 (55.5%) of the patients, 85 (39.3%) had major debridement done, and the remainder (5.2%) of the patients underwent minor debridement and medical management. Among patients carrying carbapenem-resistant gram negative pathogens, 2 patients died. In one case, the isolate corresponded K. pneumoniae and the other to A. baumannii. Both carbapenem resistant isolates harboured blaNDM-like. The Klebsiella isolate was colistin resistant as well.
DiscussionCurrently, 50.8 million Indians have diabetes. Projections indicate that India will have the largest number of diabetic patients by the year 2030AD.15 Diabetic foot infections are the most frequent complication requiring hospitalization.3,16 Microbial profiles of diabetic foot infections are widely studied and differ in different regions across India and the world. In our study, gram negative bacteria were more common isolates in DFI, similar to many studies performed in India.17,18 In contrast, gram positive organisms predominate the causatives in the western part of the world.19,20 When line listing the individual microorganisms, S. aureus was on top, followed by P. aeruginosa, similar to other studies from same region.21,22S. aureus was the most frequent pathogen (28%, 42/150), followed by P. aeruginosa (24%, 36/150) in the study by Shanthi et al.10E. coli was the most common pathogen isolated followed by S. aureus in a study from North India, but in other studies P aeruginosa has been reported most frequently.3,17 Fungal pathogens and their role in DFI is less frequently studied. A study by Chellan et al.,23 revealed that there is a high prevalence (27.9%) of fungal infection in deep tissues of diabetic lower extremity wounds with Candida parapsilosis topping the list. Fungal infections accounted for 9% of the total isolates in a study by Bansal et al.24Candida tropicalis was the most common fungal isolate in their study. In contrast, we observed lower prevalence of fungal infections in our study.
There were observed lower MRSA rates compared to other studies from India where incidence of MRSA has been reported as high as 66%.25 Our study also revealed the predominant polymicrobial nature (55.7%) of infections. Monomicrobial infections were seen only in 44.3% cases. Other studies have revealed contrasting results with monomicrobial infections being more predominant. A study by Tiwari et al.16 showed that monomicrobial infections cases (43.5%, 27/62) were more than polymicrobial infections (35.5%, 22/62). In another study, equal number of polymicrobial and monomicrobial infections were reported (44.4%).26 A study from South India found that monomicrobial infections (56%) were more predominant than polymicrobial infections (44%).27
Carbapenems are the mainstay in the treatment for multidrug-resistant gram negative bacteria; however, a rising number of carbapenemases (and thus, resistance) to this group of drugs is increasingly being reported from different parts of the world.28 Metallo-beta-lactamase have been described previously in P. aeruginosa isolates from diabetic foot infections.29 NDM is the latest in the armamentarium of carbapenemases. NDM1 (New Delhi metallo-beta-lactamase) was first identified in isolates from a Swedish patient of Indian origin in 2008.9 NDM producers have been described in studies from various parts of the world; although, many reports seem to originate from the Indian subcontinent.11,30
However, there is paucity of data with respect to prevalence of NDM among isolates from DFIs in India. Khan et al. have reported NDM producing Enterobacter cloacae and Klebsiella pneumoniae strains isolated in two patients with diabetic foot ulcers from India.31 Samant et al. have, for the first time, described Providencia rettgeri strains harbouring blaNDM-like in 4 patients with diabetic foot ulcers from India.32 In our study, we found the presence of blaNDM-like in nonfermenters like Pseudomonas and Acinetobacter as well as in E. coli and K. pneumoniae.
With the current local microbiological and antibiogram data, cefoperazone/sulbactam or piperacillin/tazobactam with clindamycin was the suggested as the empiric antibiotic of choice for DFI with escalation to carbapenem (meropenem) with teicoplanin depending upon the culture sensitivity report.
Due to resource constrains we looked only for the blaNDM-like genes. However, its worth looking into the prevalence of other carbapenemase genes like KPC, IMP, VIM etc. In one short project for CRE screening in our hospital we found an increased prevalence of KPC like genes in K. pneumoniae isolated from stool samples.(unpublished data)
ConclusionTo the best of our knowledge, this study has dealt with perhaps the largest data in the world related to diabetic foot infections for studying the pathogen spectrum along with an attempt to study the prevalence of blaNDM-like gene. This study goes on to indicate that in a tertiary diabetic foot referral centre, polymicrobial gram negative infections are more predominant in DFIs. The study has also demonstrated the emergence of difficult-to-treat gram negatives as almost one fifth of the gram negatives. Therefore, the authors believe it is now highly recommended to have strict antimicrobial stewardship programmes and restrict unnecessary use of broad spectrum antibiotics to curb the menace of antibiotic resistance.
Brief summaryAlready known:Gram negative bacteria are more common than gram positive bacteria in DFI cases in India. Monomicrobial infections are more predominant than polymicrobial infections in most studies. NDM prevalence in DFI cases has been demonstrated by few studies.13
What this study adds:We handled a larger data pool for carbapenem-resistant bacteria in DFI cases than most studies. Polymicrobial infections (54%) were found to be more predominant than monomicrobial infections (44.3%) in DFI cases. Lower rate of MRSA among gram positive bacteria. Found the presence of blaNDM-like in nonfermenters like Pseudomonas and Acinetobacter as well as in E. coli and pneumoniae.
Conflicts of interestThe authors declare no conflicts of interest.