Agricultural crops suffer many diseases, including fungal and bacterial infections, causing significant yield losses. The identification and characterisation of pathogenesis-related protein genes, such as chitinases, can lead to reduction in pathogen growth, thereby increasing tolerance against fungal pathogens. In the present study, the chitinase I gene was isolated from the genomic DNA of Barley (Hordeum vulgare L.) cultivar, Haider-93. The isolated DNA was used as template for the amplification of the ∼935bp full-length chitinase I gene. Based on the sequence of the amplified gene fragment, class I barley chitinase shares 93% amino acid sequence homology with class II wheat chitinase. Interestingly, barley class I chitinase and class II chitinase do not share sequence homology. Furthermore, the amplified fragment was expressed in Escherichia coli Rosetta strain under the control of T7 promoter in pET 30a vector. Recombinant chitinase protein of 35kDa exhibited highest expression at 0.5mM concentration of IPTG. Expressed recombinant protein of 35kDa was purified to homogeneity with affinity chromatography. Following purification, a Western blot assay for recombinant chitinase protein measuring 35kDa was developed with His-tag specific antibodies. The purified recombinant chitinase protein was demonstrated to inhibit significantly the important phytopathogenic fungi Alternaria solani, Fusarium spp, Rhizoctonia solani and Verticillium dahliae compared to the control at concentrations of 80μg and 200μg.
Agricultural crops have great economic importance worldwide, and with the population increase, the crops produced are not sufficient to feed the human population. Additionally, the agricultural crops are attacked by multiple pathogens including bacterial, viral or fungal causing reduction in both quantity and quality of the overall yield. Almost 26–30% of the yield losses for sugar beet, cotton and wheat are caused by fungal pathogens alone.1 Moreover, fungi cause 35%, 39% and 40% of the damage in maize, potato and rice, respectively.1 Fungi, such as Fusarium solani and Fusarium oxysporum, are the cause of wilting and damping off diseases, whereas Alternaria alternate causes leaf spot in various agriculturally important crops.2Rhizoctonia solani causes many diseases including black scurf in potatoes,3 root rot in sugar beet4 and sheath blight in rice.5 Similarly, Verticillium dahliae is the cause of verticillium wilt in many plants including potato,6,7 cotton,8 peppermint9 and others.
The first structure of the fungus that comes into contact with the host (plants in the case of phytopathogenic fungi) is its cell wall. Chitin, the main constituent of the fungal cell wall, is a homopolymer of N-acetylglucosamine units. In nature, plants possess natural defence systems against fungal attack. Certain plants produce antifungal proteins to defend against fungal growth.10 Isolating these antifungal proteins and inserting them into different plants increases resistance against fungal pathogens in these new host plants. These proteins include chitinases and glucanases.11 Chitinases are present in almost all organisms including bacteria,12 fungi,13 insects,14 plants15 and mammals. Chitinases act on chitin as their substrate and hydrolyse it to mono- and oligomers.16 A chitinase is classified as an endochitinase if it breaks chitin in internal sites by acting randomly; or an exochitinase, if it breaks the chitin from either nonreducing or reducing ends.17 They have different roles in different organisms but in plants chitinases are mainly involved in resistance against fungal pathogens.18 Barley possesses endochitinases (class I and class II) that can be used to control fungal pathogens.19
Escherichia coli is the most widely used bacterial expression system for producing heterologous proteins.20 To obtain a high yield of recombinant protein, the gene is usually expressed at its highest possible level. The purpose of this study was to isolate chitinase I gene from Barley and express it in an E. coli expression system to reveal its antifungal activity against four economically important phytopathogenic fungi; Alternaria solani, Fusarium spp, R. solani and V. dahliae.
Materials and methodsPlant materialBarley (Hordeum vulgare L.) variety Haider-93 grown in CEMB field was used as source material for isolation of chitinase I gene. The deduced sequence of chitinase I gene was used to obtain the amino acid sequence of the gene, which was further used for alignment and construction of a phylogenetic tree.
Gene amplification and amino acid sequence homology studiesGenomic DNA was extracted from young Barley leaves using Genomic DNA Purification kit (Thermoscientific, K0512). The 935bp chitinase I gene was amplified with 5′-AGAGCGTTCGTGTTGTTCG-3′ forward and 5′-CTGTAGCAGTCGAGGTTGTTG-3′ reverse primer in a PCR reaction mixture comprised of 2μL of 10× PCR buffer, 1.5mM MgCl2, 0.1mM dNTPs, 1pmol each of forward and reverse primers, 50ng DNA and 2 units of Taq polymerase (Thermoscientific). The PCR amplification was carried out under the following conditions: initial denaturation at 95°C for 5min, followed by 35 cycles of amplification (94°C for 45s, 61°C for 45s, and 72°C for 45s) with a final extension of 10min at 72°C. The amplified fragment was analysed on 0.8% agarose gel by electrophoresis. The amplified fragment of ∼935bp was cloned into the pCR 2.1 vector (Invitrogen, K4500-01) for sequencing. The Barley chitinase clone was sequenced through the facility at Macrogen (Korea), and edited and analysed using the BioEdit tool. The sequences of other plant chitinases used for comparison were downloaded from the National Centre of Biotechnology Information (NCBI). Multiple sequence alignment of amino acid sequences of plant chitinase retrieved from GenBank and our deduced sequence was done using Clustal W (Mega v 6) software. A phylogenetic tree was constructed using Neighbour joining method and the reliability was measured by boot strap analysis with 1500 trials using Mega v 6 software. The protein families of barley chitinase class I and class II were compared using conserved domain database, CDD, using http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi.
Construction of a prokaryote expression vector and expression of recombinant chitinaseA ∼935bp chitinase I gene was ligated in pET30a (Addgene) expression vector having N/C-terminal His-tag sequence and expressed under T7 promoter to create a pET-chiI construct. The ligation was transformed into competent cells of E. coli through the heat shock method as described by Froger and Hall.21 The pET-chiI construct DNA was mixed with 50μL of chemically competent E. coli DH5α cells and after a short incubation on ice, heat shock was given at 42°C for 45s and placed back on ice. SOC media was added and the transformed cells were incubated at 37°C for 30min with agitation.
Then, the mix was spread onto LB media plates with kanamycin for antibiotic selection and incubated at 37°C for 16–18h. Positive clones were confirmed through restriction digestion with EcoR1. For expression analyses, the pET-chiI construct was transformed in E. coli host Rosetta. E. coli Rosetta cells were transformed with the prokaryotic expression vector pET-ChiI encoding the chitinase I protein and grown in shaking culture (220rpm, 37°C) in LB supplemented with 50μg/mL kanamycin to an OD of 0.6 at 600nm. Protein expression was induced by the addition of 0.5mM isopropyl-β-d-thiogalactopyranoside (IPTG, Sigma) for 3h (220rpm 30°C). Induced cells were harvested by centrifugation at 4000×g for 10min at 4°C. The pellet was weighed, resuspended in 1× phosphate saline buffer (1:10W/V) and centrifuged at 4000×g for 10min. Lysozyme (4mg/mL) was added in the pellet and kept at room temperature for 20–30min for complete cell lysis. The lysate was sonicated (20min at 16W) and then cleared by centrifugation at 14,000×g for 15min at 4°C.
Purification was performed at 4°C inside a Kelvinator chromatography refrigerator using a 16/20 XK column with volume of 20mL and height of 15mL which was packed with Chelating Fast Flow Sepharose adsorbent (GE Healthcare; catalogue No. 17-0575-01). The column was charged with 0.2M NiCl2 solution. Binding buffer (500mM NaCl, 20mM sodium phosphate buffer, pH 7.4) was added to balance the Sepharose adsorbent. The supernatant containing the protein was filtered through 0.45μm membrane to remove cell debris. The filtered protein supernatant was then loaded onto the column at a linear flow rate of 150cm/h and the time for complete loading was calculated. Washing was performed first with wash solution 1 (20mM imidazole+binding buffer, pH 7.4) and then with wash buffer 2 (40mM imidazole+binding buffer, pH 7.4) such that all of the unbound solutes and other protein contaminants were completely removed. Elution of the His-tag recombinant chitinase I protein was done by providing step-wise increasing concentrations of imidazole (ThermoFisher Scientific; catalogue No. AC30187-0010). Five concentrations of elution buffer were prepared from 70mM to 150mM. Five fractions (1mL each) for each elution were selected and analysed by Bradford assay to check the protein concentration. At each step, fractions were analysed by SDS-PAGE on 12% polyacrylamide gel and stained with Coomassie blue (ThermoScientific). Protein size was determined using Perfect Protein™ Markers (cat #926-98000 WesternSure®). Recombinant protein was fractionated by SDS-PAGE on 12% gel using a Mini Protean II tetra cell (cat # 1658004; BioRad). The resolved purified recombinant protein was transferred to PVDF (polyvinylidenedifluoride) membrane (cat # RPN303E; Amersham) using a Trans-blot Cell (BioRad) blocked with 5% non-fat dry milk with His-tag specific antisera. Recombinant chitinase was detected with rabbit polyclonal His-tag antiserum (1: 5000 dilution; cat # sc-804; Santa Cruz). An appropriate antibody conjugated with peroxidase was used in detection with the ECL system (Amersham Biosciences).
Antifungal activity assayThe phytopathogenic fungi A. solani, Fusarium spp, R. solani and V. dahliae were provided by the Institute of Agricultural Sciences, University of the Punjab Lahore-Pakistan. Antifungal assays of purified recombinant chitinase protein were performed on the four phytopathogenic fungi by employing cylinder plate method.21 For this, three wells were made on PDA (potato dextrose agar) medium by the cylinder plate method. The PDA block containing mycelia of the test fungi was inoculated in the centre of the petri plates (23°C; 48h). When the colony size increased to 3–4cm in diameter, the wells were filled with different concentrations of purified chitinase protein; 80μg, 200μg, and control. For the control well, recombinant chitinase protein was placed in boiling water for 5min and poured into the respective well. The plates were then incubated at 23°C for 48h. Zone of inhibition created by the recombinant chitinase protein was marked and measured in comparison with negative control.
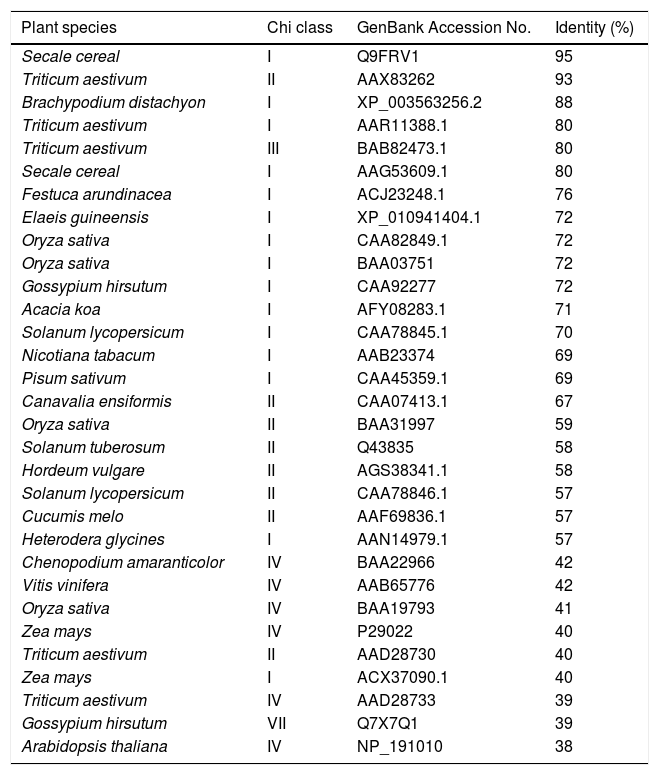
ResultsPCR amplification and amino acid sequence comparison of chitinase genesA ∼935bp chitinase I gene was successfully amplified from genomic DNA of barley using specific primers (Fig. 1A). A sharp and defined fragment can be seen in Fig. 1A. To confirm insertion, plasmid DNA was subjected to restriction digestion by EcoRI, generating two fragments; a ∼3.9kb fragment representing pCR2.1 vector and a ∼935bp fragment representing chitinase I gene (Fig. 1B). The 935bp of Chi I encoded a polypeptide of 318 amino acids. BLAST and NCBI search using the amino acid sequence indicated that it has high degree of sequence similarity with proteins encoding class I chitinase of Secale cereal, chitinases of class I, II & III of wheat (Triticum aestivum), and class I chitinase of other plants including Festuca arundinacea, Elaeis guineensis, Oryza sativa, Gossypium hirsutum, Acacia koa, Solanum lycopersicum, but has a low degree of sequence similarity with class IV chitinase of wheat (T. aestivum) and Arabidopsis thaliana, and class VII chitinase of G. hirsutum (Table 1). Moreover, it showed 58% identity with a class II chitinase of Hordeum vulgare (Fig. 2). Its highest identity is with class I chitinase of S. cereal, at 95% (Fig. 2). Wheat chitinase class II (AAX83262) does not have a chitinase binding domain and has only a catalytic site but barley class I chitinase has 93% amino acid sequence similarity to it while in comparison, barley class I chitinase has only 58% similarity to barley chitinase class II (AGS38341.1) (Supplementary Figure 1).
Amplification and cloning of chitinase I gene in TA vector and in E. coli expression vector pET30a. (A) Lane 1: 1kb DNA ladder; lanes 2–4: samples showing amplification of ∼935bp of chitinase I gene. (B) Restriction digestion of plasmid pTA:chiI harbouring chitinase I gene cloned in pCR2.1 vector with EcoRI enzyme. Lanes 1–2: pTA:chiI DNA samples; M represent 1kb DNA ladder. The release of ∼3.9kb DNA fragment represent pCR2.1 vector while ∼935bp depict chitinase gene. (C) Restriction digestion of pET-chiI recombinant plasmid with EcoRI to confirm ligation of chitinase gene in E. coli expression vector. Lane M: 1kb DNA ladder, lanes 1–3: pET-chiI plasmid DNA. The release of ∼5.4kb DNA fragment represent pET30a vector while ∼935bp depict chitinase gene.
Amino acid sequence identity of barley chitinase with other plant chitinases.
| Plant species | Chi class | GenBank Accession No. | Identity (%) |
|---|---|---|---|
| Secale cereal | I | Q9FRV1 | 95 |
| Triticum aestivum | II | AAX83262 | 93 |
| Brachypodium distachyon | I | XP_003563256.2 | 88 |
| Triticum aestivum | I | AAR11388.1 | 80 |
| Triticum aestivum | III | BAB82473.1 | 80 |
| Secale cereal | I | AAG53609.1 | 80 |
| Festuca arundinacea | I | ACJ23248.1 | 76 |
| Elaeis guineensis | I | XP_010941404.1 | 72 |
| Oryza sativa | I | CAA82849.1 | 72 |
| Oryza sativa | I | BAA03751 | 72 |
| Gossypium hirsutum | I | CAA92277 | 72 |
| Acacia koa | I | AFY08283.1 | 71 |
| Solanum lycopersicum | I | CAA78845.1 | 70 |
| Nicotiana tabacum | I | AAB23374 | 69 |
| Pisum sativum | I | CAA45359.1 | 69 |
| Canavalia ensiformis | II | CAA07413.1 | 67 |
| Oryza sativa | II | BAA31997 | 59 |
| Solanum tuberosum | II | Q43835 | 58 |
| Hordeum vulgare | II | AGS38341.1 | 58 |
| Solanum lycopersicum | II | CAA78846.1 | 57 |
| Cucumis melo | II | AAF69836.1 | 57 |
| Heterodera glycines | I | AAN14979.1 | 57 |
| Chenopodium amaranticolor | IV | BAA22966 | 42 |
| Vitis vinifera | IV | AAB65776 | 42 |
| Oryza sativa | IV | BAA19793 | 41 |
| Zea mays | IV | P29022 | 40 |
| Triticum aestivum | II | AAD28730 | 40 |
| Zea mays | I | ACX37090.1 | 40 |
| Triticum aestivum | IV | AAD28733 | 39 |
| Gossypium hirsutum | VII | Q7X7Q1 | 39 |
| Arabidopsis thaliana | IV | NP_191010 | 38 |
A ∼935bp chitinase I gene was successfully amplified from genomic DNA of barley using specific primers (Fig. 1A). A sharp and defined fragment can be seen in Fig. 1A. To confirm insertion, plasmid DNA was subjected to restriction digestion by EcoRI, generating two fragments; a ∼3.9kb fragment representing pCR2.1 vector and a ∼935bp fragment representing chitinase I gene (Fig. 1B). The 935bp of Chi I encoded a polypeptide of 318 amino acids. BLAST and NCBI search using the amino acid sequence indicated that it has high degree of sequence similarity with proteins encoding class I chitinase of Secale cereal, chitinases of class I, II & III of wheat (Triticum aestivum), and class I chitinase of other plants including Festuca arundinacea, Elaeis guineensis, Oryza sativa, Gossypium hirsutum, Acacia koa, Solanum lycopersicum, but has a low degree of sequence similarity with class IV chitinase of wheat (T. aestivum) and Arabidopsis thaliana, and class VII chitinase of G. hirsutum (Table 1). Moreover, it showed 58% identity with a class II chitinase of Hordeum vulgare (Fig. 2). Its highest identity is with class I chitinase of S. cereal, at 95% (Fig. 2). Wheat chitinase class II (AAX83262) does not have a chitinase binding domain and has only a catalytic site but barley class I chitinase has 93% amino acid sequence similarity to it while in comparison, barley class I chitinase has only 58% similarity to barley chitinase class II (AGS38341.1) (Supplementary Figure 1).
Amplification and cloning of chitinase I gene in TA vector and in E. coli expression vector pET30a. (A) Lane 1: 1kb DNA ladder; lanes 2–4: samples showing amplification of ∼935bp of chitinase I gene. (B) Restriction digestion of plasmid pTA:chiI harbouring chitinase I gene cloned in pCR2.1 vector with EcoRI enzyme. Lanes 1–2: pTA:chiI DNA samples; M represent 1kb DNA ladder. The release of ∼3.9kb DNA fragment represent pCR2.1 vector while ∼935bp depict chitinase gene. (C) Restriction digestion of pET-chiI recombinant plasmid with EcoRI to confirm ligation of chitinase gene in E. coli expression vector. Lane M: 1kb DNA ladder, lanes 1–3: pET-chiI plasmid DNA. The release of ∼5.4kb DNA fragment represent pET30a vector while ∼935bp depict chitinase gene.
Amino acid sequence identity of barley chitinase with other plant chitinases.
| Plant species | Chi class | GenBank Accession No. | Identity (%) |
|---|---|---|---|
| Secale cereal | I | Q9FRV1 | 95 |
| Triticum aestivum | II | AAX83262 | 93 |
| Brachypodium distachyon | I | XP_003563256.2 | 88 |
| Triticum aestivum | I | AAR11388.1 | 80 |
| Triticum aestivum | III | BAB82473.1 | 80 |
| Secale cereal | I | AAG53609.1 | 80 |
| Festuca arundinacea | I | ACJ23248.1 | 76 |
| Elaeis guineensis | I | XP_010941404.1 | 72 |
| Oryza sativa | I | CAA82849.1 | 72 |
| Oryza sativa | I | BAA03751 | 72 |
| Gossypium hirsutum | I | CAA92277 | 72 |
| Acacia koa | I | AFY08283.1 | 71 |
| Solanum lycopersicum | I | CAA78845.1 | 70 |
| Nicotiana tabacum | I | AAB23374 | 69 |
| Pisum sativum | I | CAA45359.1 | 69 |
| Canavalia ensiformis | II | CAA07413.1 | 67 |
| Oryza sativa | II | BAA31997 | 59 |
| Solanum tuberosum | II | Q43835 | 58 |
| Hordeum vulgare | II | AGS38341.1 | 58 |
| Solanum lycopersicum | II | CAA78846.1 | 57 |
| Cucumis melo | II | AAF69836.1 | 57 |
| Heterodera glycines | I | AAN14979.1 | 57 |
| Chenopodium amaranticolor | IV | BAA22966 | 42 |
| Vitis vinifera | IV | AAB65776 | 42 |
| Oryza sativa | IV | BAA19793 | 41 |
| Zea mays | IV | P29022 | 40 |
| Triticum aestivum | II | AAD28730 | 40 |
| Zea mays | I | ACX37090.1 | 40 |
| Triticum aestivum | IV | AAD28733 | 39 |
| Gossypium hirsutum | VII | Q7X7Q1 | 39 |
| Arabidopsis thaliana | IV | NP_191010 | 38 |
Amplified ∼935bp chitinase I gene was cloned behind T7 promoter in fusion with a His-tag in a prokaryote expression system. Recombinant clone, pET-chiI, upon restriction digestion release two size specific fragments: a ∼5.4kb fragment representing pET30a vector and a ∼935bp fragment representing the chitinase I gene (Fig. 1C). Expression of the ∼35kDa recombinant chitinase protein was optimised using SDS-PAGE and it was found that 0.5mM IPTG was best for recombinant protein expression (Fig. 3A). The expression was lowest at 0.2mM concentration of IPTG while highest at 0.5mM concentration of IPTG compared to the control, non-transformed Rosetta cells where recombinant protein is not expressed. Subsequently, the recombinant chitinase protein was purified through affinity chromatography and a thick band of ∼35kDa was observed in eluted fractions obtained at 125mM and 150mM imidazole concentration (Fig. 3B). The purified recombinant protein fractions were electrophoretically transferred onto PVDF (polyvinylidene fluoride) membrane (Millipore, USA) and incubated with polyclonal anti-His-HRP antibody (Santa Cruz, UK). An apparently homogeneous band of approximately 35kDa was detected using Chemiluminescent HRP Substrate (Millipore) (Fig. 3C).
Expression analysis of recombinant chitinase protein expressed in E. coli host Rosetta. (A) SDS-PAGE (12%) representing expression of a ∼35kDa recombinant chitinase protein optimised at various concentrations of IPTG. Lane 1: cell lysate from un-induced colony of Rosetta, lane 2: cell lysate from transformed Rosetta induced with 0.2mM IPTG, lane 3: cell lysate from transformed Rosetta induced with 0.3mM IPTG, lane 4: cell lysate from transformed Rosetta induced with 0.4mM IPTG and lane 5: cell lysate from transformed Rosetta induced with 0.5mM IPTG, lane 6: pre-stained protein marker, (B) SDS-PAGE fractionating purified fractions of recombinant chitinase protein. Lane 1: total cell lysate from un-transformed Rosetta, lanes 2–3: purified fraction of recombinant chitinase protein sample, lane 6: pre-stained protein marker, (C) Western blot of purified recombinant chitinase protein. The protein was detected through Histag specific antibodies using ECL system. Lanes 1–2: purified fraction of 35kDa chitinase protein.
Purified recombinant chitinase protein, measuring ∼35kDa when applied on four important phytopathogenic fungi, exhibited strong inhibitory effect on the mycelial growth of the subjected fungi. Significant growth retardation in the mycelial growth of A. solani, Fusarium spp and V. dahliae was observed at 200μg concentration of purified chitinase protein while the zone of inhibition was less around the wells where 80μg of the purified protein was applied. In the control sample, no inhibitory effect was measured (Fig. 4). In R. solani, less inhibition was observed at 80μg concentration of the purified protein compared to other tested fungi while at 200g concentration, a zone of inhibition can be seen (Fig. 4). It can be concluded that recombinant chitinase protein, when over-expressed exhibits strong and significant antifungal potential. Hyphal growth of the inoculated fungi was restricted to a particular zone of inhibition.
Antifungal activity assay of purified recombinant chitinase protein towards (A) Alternaria solani, (B) Fusarium spp., (C) Verticillium dahliae, (D) Rhizoctonia solani. Each selected fungus was subjected to two protein treatments, 80μg and 200μg along with control. The letter ‘C’ represent control that contains heat deactivated purified chitinase protein.
Chitinases belong to group of pathogenesis-related (PR) proteins in plants22–25 and several of them have been employed to enhance the tolerance of the plant towards a particular pathogen through genetic transformation technologies.26,27 Our aim was to isolate a plant based chitinase I gene, over express it into a prokaryote expression vector under T7 promoter and reveal the antifungal potential of the purified recombinant protein against four important phytopathogenic fungi.
In our study, we selected a plant chitinase as they are usually endo-chitinases and contain lysozyme activity. While, in contrast, the bacterial chitinases are exo-chitinases and hydrolyse the chromogenic trisaccharide analogue pNPP (p-nitrophenyl-P-D-N, N′-diacetylchitobiose) as the substrate. Additionally, plant chitinase I is reported to exhibit high antifungal activity compared to other plant chitinases.28 Barley-derived chitinase genes have been used to enhance fungal resistance in many crops like brassica,19 wheat,29 rice30 and carrot.31 The chitinase I gene is evolutionarily conserved and is present in both monocot and dicot plants such as cocoa,32 potato33 and rice.34 The chitinase I gene also has a chitin binding domain, which is absent in class II chitinase and hence make it superior in antifungal potential.
We documented an interesting finding that barley-derived chitinase I gene and II gene not only differs in presence of chitin binding domain but also do not share sequence homology. Rather, barley chitinase class I shares 93% amino acid sequence homology with chitinase class II of wheat. Furthermore, we employed pET expression system for expression studies of chitinase recombinant protein as previously done by Liu and Naismith.35 This is the most widely used system for the cloning and in vivo expression of recombinant proteins in E. coli. The reason behind this is the high selectivity of the pET system's bacteriophage T7 RNA polymerase for its cognate promoter sequences, the high level of activity of the polymerase and the high translation efficiency mediated by the T7 gene 10 translation initiation signals.36 We employed IPTG as an inducing agent as it is reported to provide better induction.37
Our antifungal result showed that 80μg of recombinant chitinase protein has a clear zone of inhibition while 200μg of the protein has an increased zone of inhibition. There are reports38 that rice chitinase had no significant effect on fungi tested at 100μg and had only subtle effect at a higher dose of 300μg. Kirubakaran and Sakthivel used E. coli strain BL-21 as host and pET 28a+ vector for the overexpression of chitinase I gene and analysed its antifungal activity against different fungal pathogens and reported that 300μg of enzyme activity has broad spectrum anti-fungal inhibition.22 This finding demonstrates that our chitinase isolate has much better inhibitory effect on test fungi at low doses than isolates used in other studies.
In conclusion, Chitinases that cleave chitin polymers in the fungal cell wall are used to increase resistance to fungal pathogens. And for this purpose, adequate antifungal activity of the chitinase is mandatory. Therefore, screening chitinase with high antifungal activity is important to meet the need to improve resistance against phytopathogens by gene manipulation. Attempts have been made by several researchers to identify new chitinase genes with enhanced antifungal potential. Like Liu et al. expressed two chitinase genes, LbCHI31 and LbCHI32 from Limonium bicolor in E. coli BL21 strain and it has significant inhibition against A. alternate.39 In a similar study, Itoh et al. overexpressed a cell surface-expressed chitinase from Paenibacillus in E. coli.40 In another study, horsetail derived chitinase gene was overproduced in E. coli.41 In a similar study, a chitinase gene (pcht28), isolated from Lycopersicon chilense was transformed to strawberry and its expression was checked against V. dahliae. It showed more resistance against these fungi compared to control, untransformed. This suggests pcht28 plays a role in defence against fungal disease caused by V. dahliae.42 Mishra et al. transformed the Trichoderma endochitinase gene into guava and measured its inhibitory action against F. oxysporum, the cause of fusarium wilt.43 In our findings, overexpression of chitinase I gene results in elevated chitinolytic activity, induction of expression of the recombinant chitinase gene and increased inhibition for fungal phytopathogens. All these results collectively indicate that the chitinase I gene derived from barley can be useful in gene engineering against plant diseases.
Conflicts of interestThe authors declare no conflicts of interest.