The biodegradation of synthetic dyes by fungi is emerging as an effective and promising approach. In the present study, freshwater fungal strains isolated from submerged woods were screened for the decolorization of 7 synthetic dyes. Subsequently, 13 isolates with high decolorization capability were assessed in a liquid system; they belonged to 9 different fungal species. Several strains exhibited a highly effective decolorization of multiple types of dyes. New absorbance peaks appeared after the treatment with 3 fungal strains, which suggests that a biotransformation process occurred through fungal biodegradation. These results showed the unexploited and valuable capability of freshwater fungi for the treatment of dye-containing effluents. The ability of certain fungi to decolorize dyes is reported here for the first time.
Synthetic dyes are widely used for coloring the products of several industries such as textiles, leather, cosmetics, paper, printing materials, and plastics. It is estimated that 1–2% of dye production is lost, and 5–10% is discharged to the environment when the dyes are used.1,2 Several dyes and the chemicals used to produce them are often toxic, carcinogenic or even explosive.3,4 Effluents from industries that use various dyes are considered as pollutants that can cause severe environmental problems as well as medical and esthetic problems.2 The decolorization of this industrial waste is a challenging task because certain dyes are resistant to degradation.4
Physical and chemical decolorizing methods have been developed to remove dyes from wastewater; however, several of them have disadvantages such as high costs and/or limited applicability.2,5 Studies on the capability of microorganisms to perform dye decolorization has received increasing attention because the use of microorganisms is considered a cost effective and environmentally friendly method for removing organic dyes from wastewater before they are discharged.6,7 Likewise, the capabilities and mechanisms of decolorization by bacteria have been studied.8,9 However, the application of bacteria was limited due to the narrow substrate range of various degrading bacteria.10 Moreover, to complete the degradation of dyes, different groups of bacteria are required in an alternation process from anaerobic to aerobic conditions.2,7
Research on the fungal decolorization of dye wastewater has been performed in recent years.11,12 Several fungi with the capability to decolorize a wide range of dyes have been reported. For example, the white-rot fungi and brown-rot fungi are well-studied fungi groups with decolorization abilities.12 Other fungi, such as Aspergillus niger, Rhizopus arrhizus, and R. oryzae, can also decolorize and/or absorb diverse dyes and possess excellent color removal capabilities.13,14
The mechanism of fungal decolorization mainly involves two aspects, biodegradation and biosorption.15 The biodegradation capability of fungi is due to their extracellular, non-specific and non-selective enzyme system.6 Fungal enzyme production depends on nutrient limitations, and their subsequent dye decolorization ability is achieved depending on the growth conditions.16 Considering the complex environmental factors involved in the dye wastewater conditions, the screening of more fungi is necessary for use in dye decolorization.
Aquatic fungi are the main decomposers of aquatic ecosystems and play crucial roles in the cycling of nutrients.17 In addition, a unique characteristic of fungi is their ability to produce several non-specific enzymes.18 These non-specific extracellular and/or exoenzymes enable the aquatic fungi to attack structurally diverse organic compounds that correspond to different pollutant classes.19,20 Hence, these fungi may serve as a new resource to treat wastewater. However, little research has been performed on the process of decolorizing wastewater by freshwater fungi,10 and more fungi with potential for the biodegradation of synthetic dyes need to be explored. In the present study, several freshwater fungi were isolated from streams in the Zhejiang Province, PR China, and their ability of decolorizing multiple synthetic dyes was evaluated.
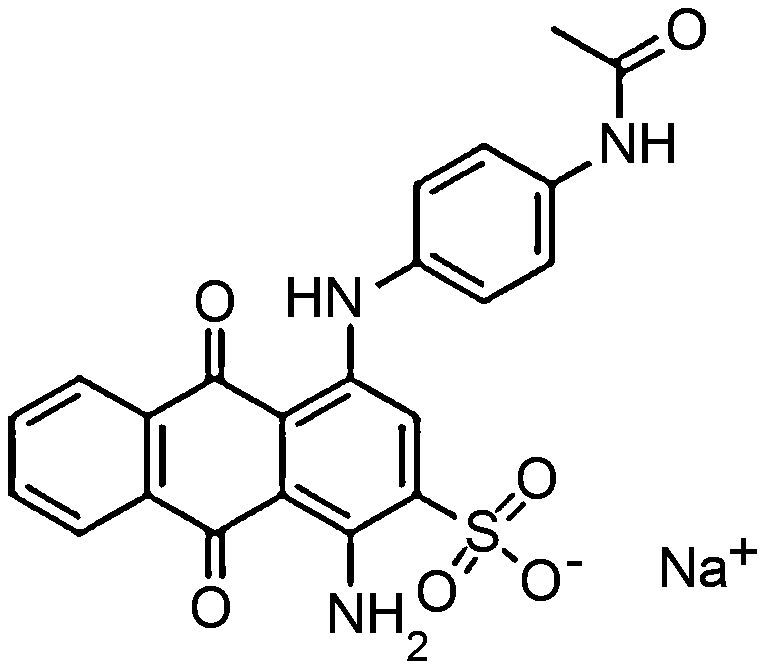
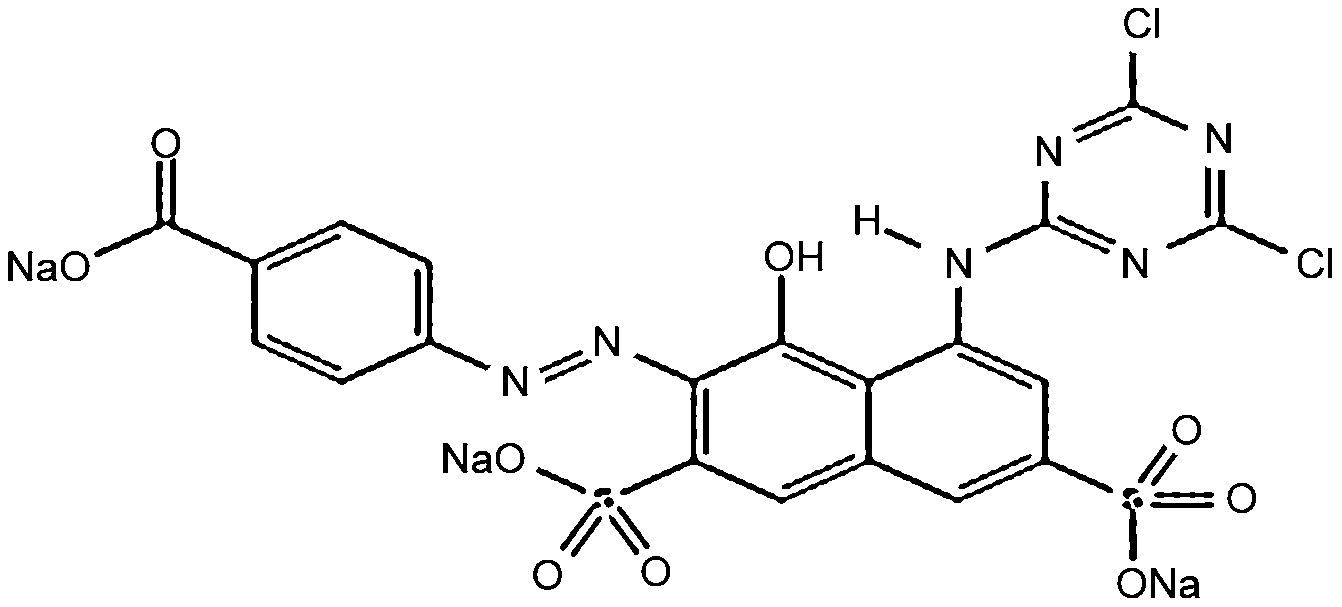
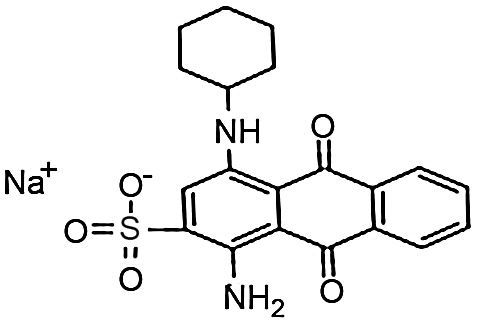
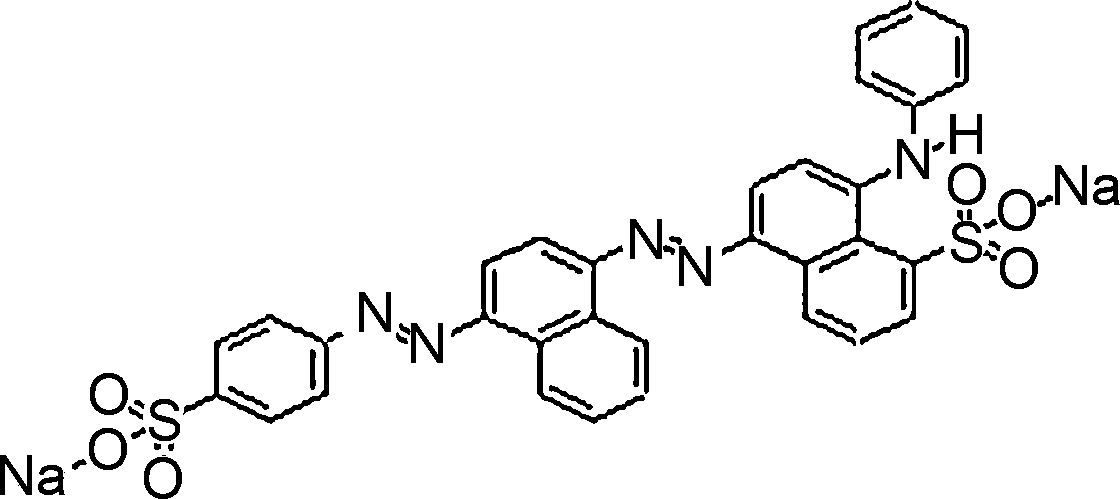
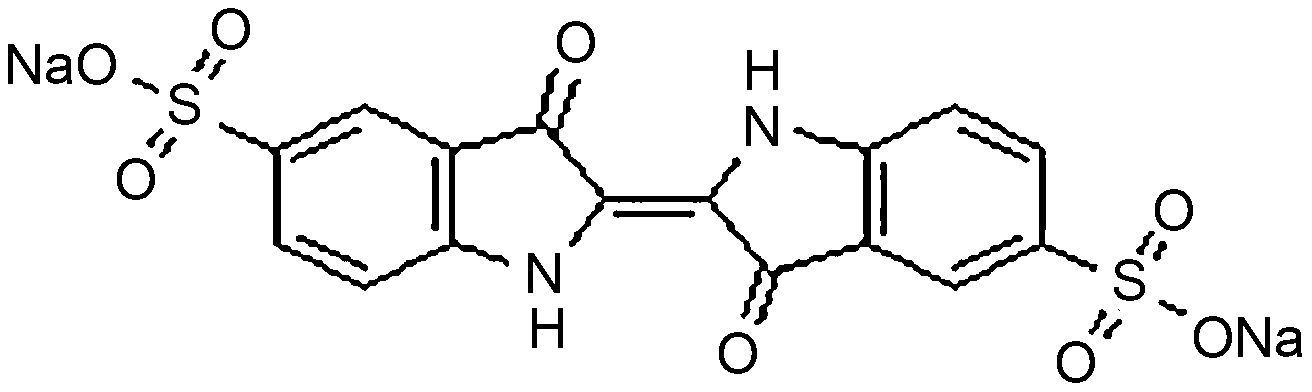
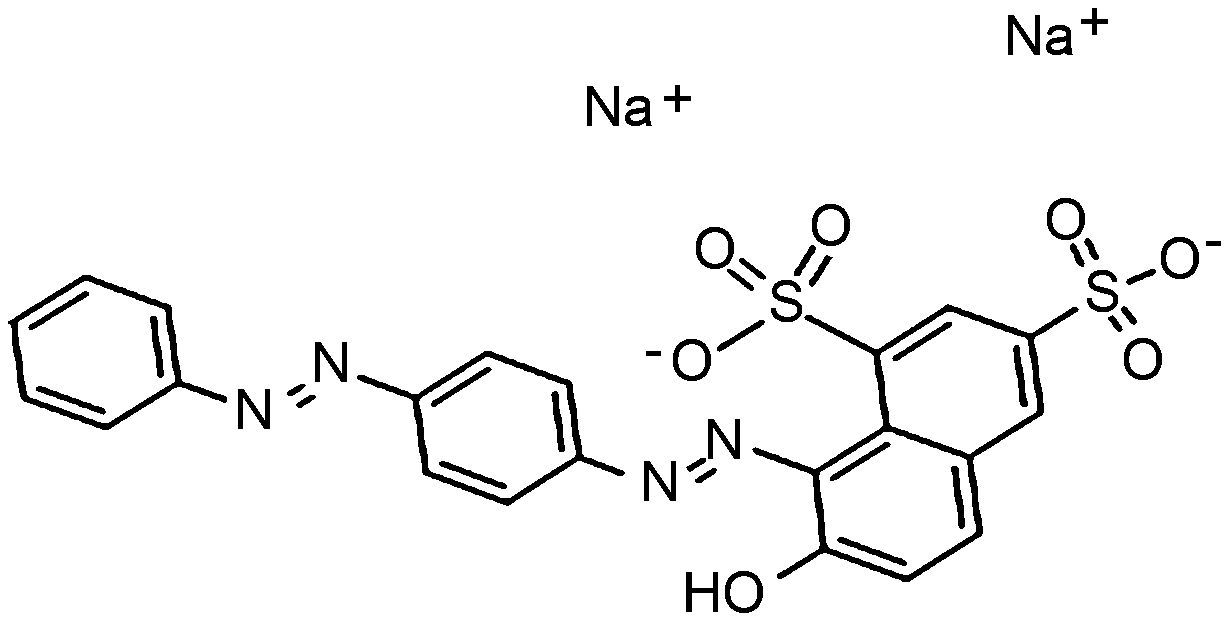
Materials and methodsDyes and mediaSeven dyes were used for screening the decolorization ability of the fungal isolates. All the dyes were dissolved in distilled water at a concentration of 10g/L; then, they were filtered and sterilized using a 0.22μm-diameter bacteria filter before being tested for use. All the dyes were purchased from Sigma–Aldrich. The molecular structures of the dyes are listed in Table 1.
Water agar medium (WA; WA1.5% agar) was used for the fungal isolation; potato-carrot-agar medium (PCA; 5% potato, 5% carrot, 2% agar) was used for the identification of fungal isolates; potato-dextrose-agar medium (PDA; 20% potato, 2% dextrose, 2% agar) was used to grow the fungal cultures and for the DNA extraction; and malt-agar medium (MEA; 2% malt extract, 1.5% agar) was used for screening the decolorization by fungi.
The liquid MEA medium used for the decolorization test was the same as that described above but without agar. The PDA medium was used for colony growth, and the genomic DNA isolation was prepared following the method of Wongsawas et al.21
Fungal strain isolation from streamsSubmerged wood samples were randomly collected from streams in the Zhejiang Province; then, they were brought back to laboratory and placed separately in snap lock plastic bags with sterile moist paper towels, incubated at room temperature and examined periodically during a three-month period. Single spore isolations were obtained using the methods described by Choi et al.22,23
Dye decolorization on solid mediaThe solid media were prepared with MEA medium and the addition of each dye to a total concentration of 50mg/L. A mycelium plug derived from the edge of fungal strains grown on WA medium plates for 4 days at 25°C were transferred to the center of a solid medium plate and inoculated at 25°C for 1 week. The formation of decolorized zones under or around the developing mycelia was monitored for dye decolorization. All the agar plate assays were performed in triplicate.
Morphological identification of fungal strains with decolorization capabilityPure cultures of fungi with decolorization capabilities were incubated on PCA, WA and PDA media at 25°C for 2 weeks for their identification. The morphological characters of fungi were examined and recorded in detail under an Olympus BX-51 microscope equipped with an Olympus DP-50 digital camera system.
Molecular identification of the fungal strains testedThe total genomic DNA was extracted from the cultured mycelia with a modified CTAB method protocol.21 The complete internal transcribed spacers 1 and 2 and the 5.8S rDNA regions of the isolates tested were amplified by PCR using ITS1 and ITS4 primers. The 18S rDNA region of isolate JX-43 was obtained by PCR using the NS1 and NS4 primers because there is no ITS sequence of this species in GenBank. The PCR reaction protocol and sequencing process was performed as described by Wongsawas et al.21
Dye decolorization in liquid systemsThe fungal strains that showed a decolorized zone on the solid media were picked out for further testing in liquid media. The concentration of each dye was supplemented to obtain 50mg/L in a previously autoclaved liquid medium. A plug derived from the edge of the selected fungal strains on PDA was transferred to a 10mL liquid medium that contained the respective dye and inoculated at 25°C for 1 week. Then, the liquid medium that contained fungal mycelia was centrifuged at 5000rpm for 10min, and aliquots of the supernatants were transferred into new tubes and diluted as necessary for the absorbance readings. For each dye, untreated liquid medium was used as a control. The absorbance spectrum of each dye was examined using a U-2001 two-beam spectrophotometer using a wavelength range from 200 to 900nm, and the maximum absorbance wavelength was determined according to the single peak with the strongest absorption. The absorbance determinations were measured at the maximum absorbance wavelength on a GENios+microplate reader. The standard curves of each dye were performed at the maximum absorbance wavelength, and the dye concentration ranged from 0 to 50mg/L. The dye decolorization of samples was expressed as a percentage, as follows: D=(AC−AA)/AC×100, where D is the decolorization rate (%), and AC and AA are the concentrations measured for the control and the treated sample, respectively.24
The dye biosorption of fungal mycelia was evaluated as follows: after the centrifugal process described above, the fungal mycelia in each centrifuge tube were put on a filter paper (Whatman) to remove the additional liquid; then, they were transferred to a new centrifuge tube that contained 10mL ethanol and 10mL fresh liquid medium. These tubes were placed on a rotary shaker (160rpm) at room temperature for 20min and then centrifuged at 5000rpm for 10min. The control for each dye was performed by adding 10mL ethanol into 10mL liquid medium that contained 50mg/L of each dye. The dye biosorption rate was expressed as a percentage, as follows: A=AM/AD×100, where A is the dye adsorption rate of each fungal mycelia (%), and AM and AD are the concentrations measured in the treatment and control, respectively.
The assays were performed in triplicate, the experimental error was calculated as the standard deviation (SD) and, in all the cases, the SD was below 3%.
ResultsFreshwater fungal strain isolation and screening of the dye decolorization ability on agar platesA total of 92 strains of freshwater fungi were isolated from decayed woody samples collected from streams in the Zhejiang Province, PR China. The decolorization capability of these fungal strains was assessed on dye-containing MEA plates with 7 types of synthetic dyes inoculated at 25°C for 1 week.
Thirteen fungal strains that caused either a partial or full decolorization were screened out of 92 strains of freshwater fungi isolated from the decayed wood samples. The dye decolorization results on agar plates showed that these fungal strains had the ability to decompose more than 3 types of dyes. Likewise, different fungi had different dye decolorization abilities. For example, strain C-1 had the capability to decompose all 7 different types of dyes tested in these experiments (Fig. 1).
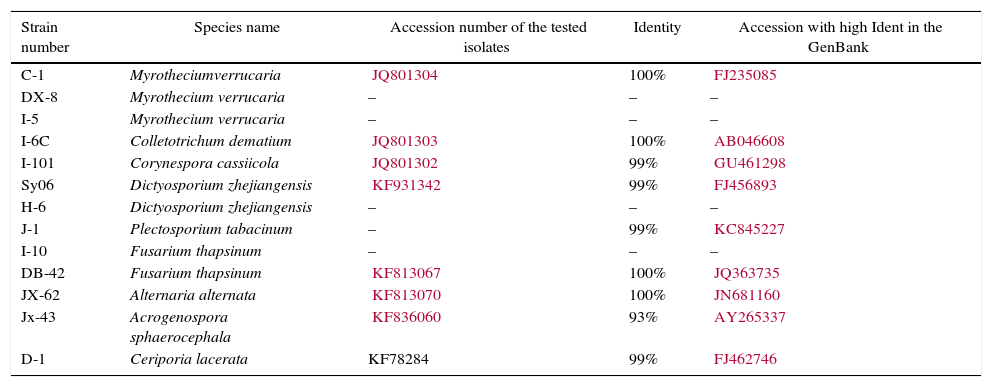
Identification of the fungi with potential for dye decompositionThe identification of the 13 fungal strains was performed based on morphological characters and on the ITS rDNA sequence (Table 2). Based on the recorded morphological data (not shown) from 3 strains (DX-8, I-5 and C-1), it appears that they belong to the species Myrothecium verrucaria, with slightly different cultural characteristics (Fig. 2) and different abilities to decompose dyes (Table 3). The ITS regions of strain C-1 were sequenced, and a preliminary sequencing analysis confirmed that the taxon belonged to the Myrothecium species. The fungal strain I-6c was identified as Colletotrichum dematium. The fungal strain I-101 was similar to Corynespora cassiicola, while strains H-6 and Sy06 possessed morphology similar to that of Dictyosporium zhejiangensis. The morphology of I-10 and BD-42 were consistent with that of Fusarium thapsinum.25 The fungal strain JX-62 had the morphological characters of Alternaria alternata.
Fungal strains used for the decolorization assay after the screening test and their blast results obtained from GenBank.
| Strain number | Species name | Accession number of the tested isolates | Identity | Accession with high Ident in the GenBank |
|---|---|---|---|---|
| C-1 | Myrotheciumverrucaria | JQ801304 | 100% | FJ235085 |
| DX-8 | Myrothecium verrucaria | – | – | – |
| I-5 | Myrothecium verrucaria | – | – | – |
| I-6C | Colletotrichum dematium | JQ801303 | 100% | AB046608 |
| I-101 | Corynespora cassiicola | JQ801302 | 99% | GU461298 |
| Sy06 | Dictyosporium zhejiangensis | KF931342 | 99% | FJ456893 |
| H-6 | Dictyosporium zhejiangensis | – | – | – |
| J-1 | Plectosporium tabacinum | – | 99% | KC845227 |
| I-10 | Fusarium thapsinum | – | – | – |
| DB-42 | Fusarium thapsinum | KF813067 | 100% | JQ363735 |
| JX-62 | Alternaria alternata | KF813070 | 100% | JN681160 |
| Jx-43 | Acrogenospora sphaerocephala | KF836060 | 93% | AY265337 |
| D-1 | Ceriporia lacerata | KF78284 | 99% | FJ462746 |
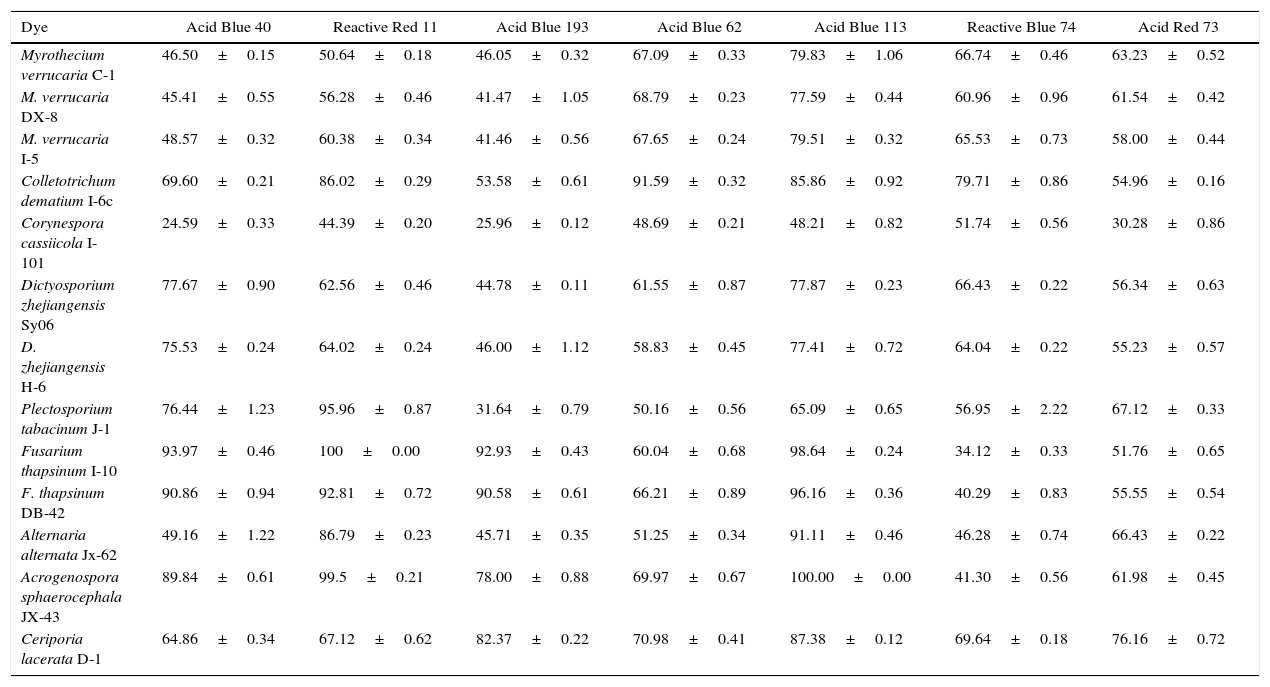
Decolorization rates (%) of 11 dyes after the treatment with 13 fungal strains for 7 days in liquid media.
| Dye | Acid Blue 40 | Reactive Red 11 | Acid Blue 193 | Acid Blue 62 | Acid Blue 113 | Reactive Blue 74 | Acid Red 73 |
|---|---|---|---|---|---|---|---|
| Myrothecium verrucaria C-1 | 46.50±0.15 | 50.64±0.18 | 46.05±0.32 | 67.09±0.33 | 79.83±1.06 | 66.74±0.46 | 63.23±0.52 |
| M. verrucaria DX-8 | 45.41±0.55 | 56.28±0.46 | 41.47±1.05 | 68.79±0.23 | 77.59±0.44 | 60.96±0.96 | 61.54±0.42 |
| M. verrucaria I-5 | 48.57±0.32 | 60.38±0.34 | 41.46±0.56 | 67.65±0.24 | 79.51±0.32 | 65.53±0.73 | 58.00±0.44 |
| Colletotrichum dematium I-6c | 69.60±0.21 | 86.02±0.29 | 53.58±0.61 | 91.59±0.32 | 85.86±0.92 | 79.71±0.86 | 54.96±0.16 |
| Corynespora cassiicola I-101 | 24.59±0.33 | 44.39±0.20 | 25.96±0.12 | 48.69±0.21 | 48.21±0.82 | 51.74±0.56 | 30.28±0.86 |
| Dictyosporium zhejiangensis Sy06 | 77.67±0.90 | 62.56±0.46 | 44.78±0.11 | 61.55±0.87 | 77.87±0.23 | 66.43±0.22 | 56.34±0.63 |
| D. zhejiangensis H-6 | 75.53±0.24 | 64.02±0.24 | 46.00±1.12 | 58.83±0.45 | 77.41±0.72 | 64.04±0.22 | 55.23±0.57 |
| Plectosporium tabacinum J-1 | 76.44±1.23 | 95.96±0.87 | 31.64±0.79 | 50.16±0.56 | 65.09±0.65 | 56.95±2.22 | 67.12±0.33 |
| Fusarium thapsinum I-10 | 93.97±0.46 | 100±0.00 | 92.93±0.43 | 60.04±0.68 | 98.64±0.24 | 34.12±0.33 | 51.76±0.65 |
| F. thapsinum DB-42 | 90.86±0.94 | 92.81±0.72 | 90.58±0.61 | 66.21±0.89 | 96.16±0.36 | 40.29±0.83 | 55.55±0.54 |
| Alternaria alternata Jx-62 | 49.16±1.22 | 86.79±0.23 | 45.71±0.35 | 51.25±0.34 | 91.11±0.46 | 46.28±0.74 | 66.43±0.22 |
| Acrogenospora sphaerocephala JX-43 | 89.84±0.61 | 99.5±0.21 | 78.00±0.88 | 69.97±0.67 | 100.00±0.00 | 41.30±0.56 | 61.98±0.45 |
| Ceriporia lacerata D-1 | 64.86±0.34 | 67.12±0.62 | 82.37±0.22 | 70.98±0.41 | 87.38±0.12 | 69.64±0.18 | 76.16±0.72 |
The morphological characteristics of J-1 indicate a close resemblance to Plectosporium tabacinum. The ITS sequences further confirmed such an affinity. Strain JX-43 was similar to Acrogenospora sphaerocephala; however, the ITS sequence of this strain did not reveal high sequence similarities because no ITS sequence data of this species were found in the GenBank database. Then, we sequenced the SSU sequence of strain JX-43, and a blast search result indicated that it had 99% identity with Farlowiella carmichaeliana, the teleomorph of A. sphaerocephala. The D-1 strain could not be identified based on morphology because it was unable to sporulate; however, the ITS sequences show a close similarity to Ceriporia lacerate, known as the white-rot fungus.26
All 7 types of dyes were used for the decolorization test in liquid media. The wavelengths of maximum absorption by each dye are listed in Table 1. The 13 fungal strains that were significantly active in dye decolorization in solid plates were used in the liquid decolorization system assay. All the tested strains showed a highly efficient decolorization of a variety of dyes (Table 3). The decolorizing extent of the dyes by the different strains exhibited an individual pattern based on the dye type and the fungal strains, even those that belonged to the same fungal species. Each strain had the ability of decolorize multiple dyes and, at least, they decolorized one dye with high efficiency. For example, strain D-1 could decolorize all the tested dyes with a rate of up to 50%. Strains JX-43 and I-10 were able to decolorize the Reactive Red 11 and Acid Blue 113 dyes almost completely; however, only 34.1% and 41.3% decolorization rates were achieved with Reactive Blue 74, respectively. Certain isolates of this study had a decolorization efficiency higher than that of the white-rot fungus Ceriporia lacerata. The obtained results are summarized in Table 3.
New absorbance peaks appeared after the treatment of dyes with the fungal strains. A new absorbance peak was shown when Acid Blue 62 was treated with sy06 and I-6C respectively, which has a maximum absorbance wavelength at 590nm (Fig. 3).
In this study, the dye biosorption by the fungal strains was evaluated (Table 4). Different strains exhibited different extents of dye absorbance depending on the type of dyes. Strain I-101 had the ability of adsorbing 6 out of 7 dyes. In addition, 8 out of 13 strains were able to adsorb Acid Red 73, whereas the dye Acid Blue 193 exhibited a null adsorption by all the fungal strains used in this study. Strain I-101 has the highest rate of biosorption among the tested fungal strains, and the efficiency of dye biosorption was more than 20% in 5 types of dyes. However, at least 4 strains, C-1, DX-8, I-5, DB-42, did not show any biosorption ability.
Biosorption rates (%) of 11 dyes after the treatment with 13 fungal strains for 7 days in liquid media.
| Dye | Acid Blue 40 | Reactive Red 11 | Acid Blue 193 | Acid Blue 62 | Acid Blue 113 | Reactive Blue 74 | Acid Red 73 |
|---|---|---|---|---|---|---|---|
| Myrothecium verrucaria C-1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| M. verrucaria DX-8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| M. verrucaria I-5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Colletotrichum dematium I-6c | 8.58±0.93 | 0 | 0 | 26.50±0.64 | 1.40±0.32 | 2.36±0.35 | 14.03±0.58 |
| Corynespora cassiicola I-101 | 20.10±0.36 | 29.01±1.41 | 0 | 30.10±0.73 | 26.17±0.85 | 20.87±0.88 | 1.12±0.12 |
| Dictyosporium zhejiangensis Sy06 | 0 | 0 | 0 | 0 | 0 | 0 | 32.0±0.99 |
| D. zhejiangensis H-6 | 0 | 0 | 0 | 0 | 0 | 0 | 41.84±2.41 |
| Plectosporium tabacinum J-1 | 0 | 0 | 0 | 0 | 0 | 0 | 10.03±1.88 |
| Fusarium thapsinum I-10 | 0 | 0 | 0 | 0 | 0 | 0 | 8.03±2.46 |
| F. thapsinum DB-42 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Alternaria alternata Jx-62 | 2.22±0.44 | 0 | 0 | 0 | 0 | 0 | 24.44±1.53 |
| Acrogenospora sphaerocephala JX-43 | 0 | 0 | 0 | 0 | 0 | 0 | 12.03±0.66 |
| Ceriporia lacerata D-1 | 0 | 0 | 0 | 4.78±0.88 | 0 | 0 | 0 |
The presence of dyes in industrial products currently results in wastewater pollution, and these pollutants need to be treated before they are discharged.27 The ability of microorganisms to perform dye degradation has received considerable attention.7,16 The effluents from the textile industry are extremely variable in composition; even a small structural difference can affect the decolorization process.2,28 There is still a need for studies to discover the potential of fungal isolates for the improvement of dye wastewater because certain isolates cannot be suitable for a variety of dyes in wastewater conditions.1 The fungal isolates in this research provide a new avenue for approaching the biodegradation of dyes in the future.
The fungi used for dye biodegradation were mainly isolated from forest ecosystems, such as white-rot and brown-rot fungi. Although these fungi have been shown to decolorize dyes in liquid fermentations, their enzyme production was shown to be unreliable. This outcome is mainly due to the unfamiliar environment of liquid fermentations for these forest fungi.12 New isolates of fungi used to decolorize water conditions will probably have the efficacy of bioremediation of dyes because these fungi have the ability to secrete extracellular non-specific enzymes that contribute to the carbon recycling in liquid environments. Junghanns et al.10 showed that certain isolates of aquatic fungi had a high and unexploited potential for the treatment of dye-containing wastewaters, even at high concentrations. In the present study, 13 isolates of freshwater fungi showed a high efficiency for the decolorization of multiple dyes, which should provide advantages for the treatment of effluents with complex dye compositions. The decolorization abilities of several fungal species in decolorizing synthetic dyes, such as A. sphaerocephala, D. zhejiangensis, and P. tabacinum, have never been reported before.
The mechanisms of fungal dye removal are categorized into the following three groups: bioaccumulation, bioabsorption and biodegradation.7,16 Bioaccumulation is referred to the accumulation of pollutants by actively growing cells through their metabolism.16 Biosorption can occur in either living or dead biomass; amino, carboxyl, thiol, lipid and phosphate groups that exist in the fungal cell walls are responsible for binding the dye molecules in the biosorption process.16,29 The main mechanism of decolorization by fungi is biodegradation because the fungi can produce various non-specific extracellular and intracellular enzymes involved in the dye decolorization process, such as laccase, manganese peroxidase (MnP), manganese independent peroxidase (MIP), lignin peroxidase (LiP), tyrosinase, and others.1,16,29 The biodegradation ability of fungi through enzymatic activities was confirmed by using purified enzymes for decolorization.11,16 Moreover, there are several factors that can influence the rate of fungal dye biodegradation, such as environmental parameters, the dye molecule structures, and the relative contributions of enzymes involved in the decolorization, which may be different for each fungus.1,16,30 Further research is needed to understand the factors that influence the decolorization rate with the studied fungi.
Studies on the bio-decolorization of textile dye effluents by fungi have been reported even in the presence of salts and high dye concentration using the white-rot fungi,31Aspergillus fumigatus and Phanerochaete sordid.28,32 In the present study, one isolate, D-1, which was identified as the white-rot fungus C. lacerate,26 also showed a powerful ability to decolorize multiple dyes. Certain fungal isolates in this research, such as C. dematium strain I-6c, F. thapsinum strain DB-42, F. thapsinum strain I-10, and A. sphaerocephala strain JX-43, showed a higher potential for dye decolorization than the D-1 strain, which indicates their suitability for the treatment of dye-polluted water. The assessment of the removal abilities of multiple dyes by certain fungal strains indicates that several enzymes were involved in the decolorization process although the enzymes contributing to dye decolourisation remain to be defined, for the dye biosorption contributing to decolorisation by mycelia was very low. Moreover, the high frequency appearance of new absorbance peaks after fungal treatment demonstrated the roles of the enzymes involved in both the decomposition and the biotransformation of dyes. This characterization of fungi revealed that they may be useful for the treatment of dye-containing wastewaters.
Conflicts of interestThe authors declare no conflicts of interest.
This research was supported by the Major Science & Technology Project of the Xinjiang Uygur Autonomous Region (no. 201130102) and the Natural Science Foundation of the Zhejiang Province (LY15C140001).