To investigate the biological behavior of classical and atypical osteoblastomas in comparison to osteosarcomas.
METHODS:Based on histological parameters, 30 osteoblastomas were subclassified as classical osteoblastomas (23/30) or atypical osteoblastoma (high cellularity, prominent blue osteoid, and epithelioid osteoblasts–7/30). Comparative immunohistochemical and clinical analysis was performed in 17 cases of patients with high-grade osteosarcoma. Formalin-fixed, paraffin-embedded archival tissue was immunostained for p53 and proliferation cell nuclear antigen. Tumors with positive p53 stain underwent molecular analyses for fragments of exon 10.
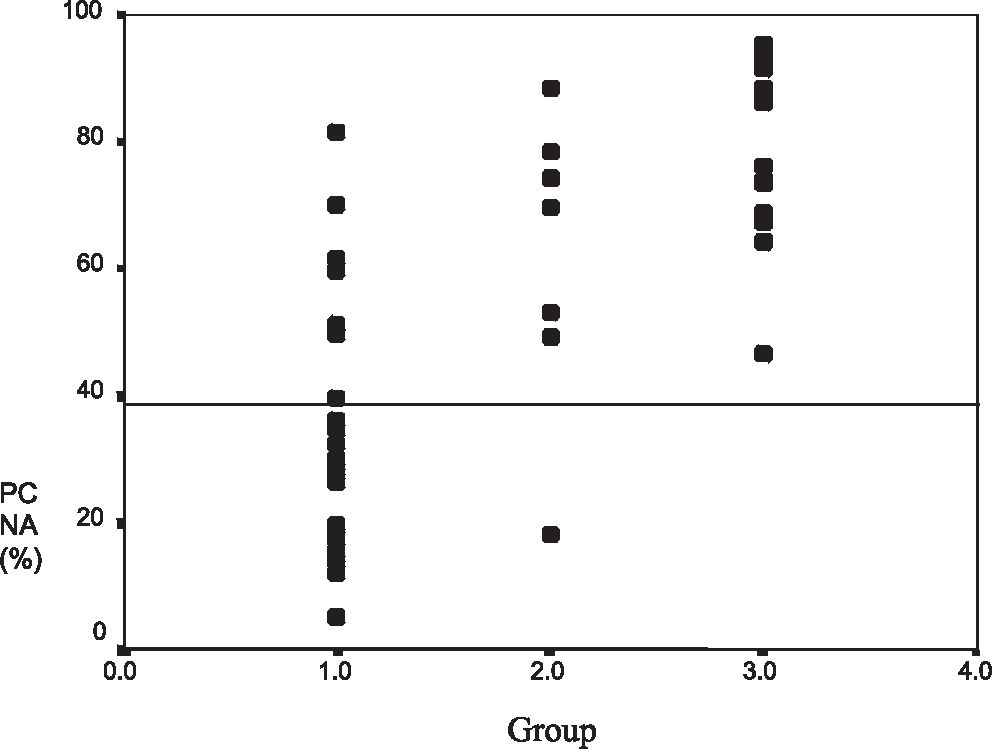
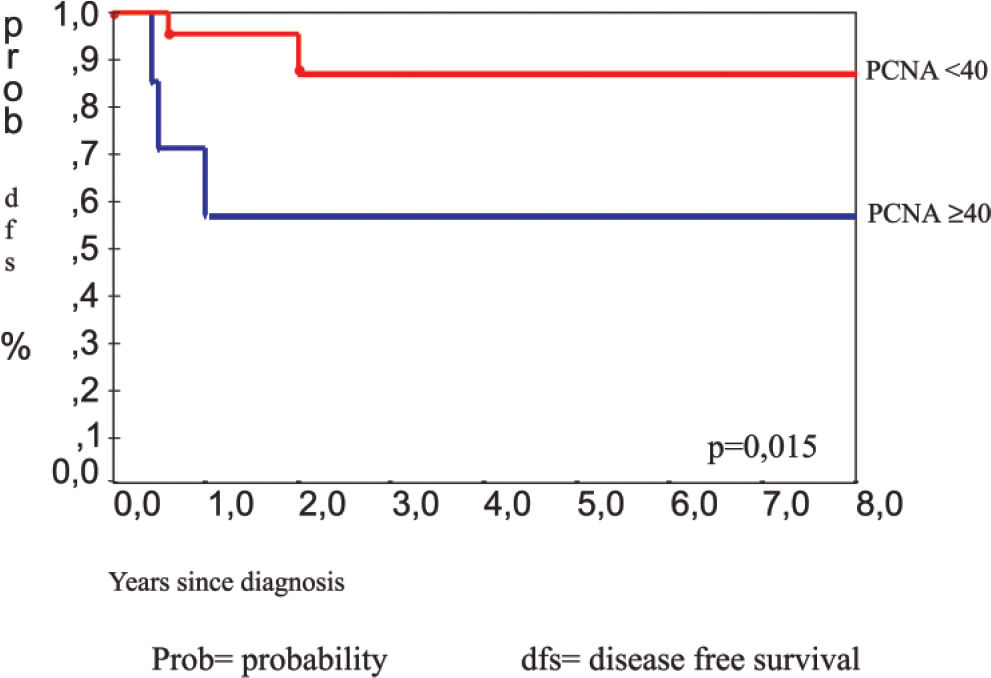
RESULTS:The mean proliferating cell nuclear antigen indexes for classical osteoblastoma, atypical osteoblastoma, and osteosarcoma were 33%, 61%, and 79%, respectively. The atypical subgroup showed similar results to those of the osteosarcoma group (P < 0.001). p53 protein was detected in 4 (13%) osteoblastomas (3 of these were atypical osteoblastoma), and 4 osteosarcomas (23%) also showed p53 positivity. DNA mutation performed in p53-positive cases was confirmed in exon 10 in 2 atypical osteoblastomas (2/3), 1 classical osteoblastoma (1/1), and 1 osteosarcoma (1/4). Disease recurrence was correlated with p53 expression (P = 0.009), atypical subtype (P = 0.031), spiculated blue bone on histology (P = 0.018), and proliferating cell nuclear antigen labeling index ≥ 40 (P = 0.015).
CONCLUSION:These results validate atypical osteoblastoma as an entity, with p53 and proliferation cell nuclear antigen immunoexpression closer to that of osteosarcoma than of classical osteoblastoma. Proliferating cell nuclear antigen labeling index and p53 may be useful predictors of recurrence.
Investigar o comportamento biológico de osteoblastomas clássicos e atípicos comparados com osteossarcomas.
MÉTODOS:Com base em parâmetros histológicos classificamos um grupo de 30 osteoblastomas nos subgrupos de osteoblastomas clássicos (23/30) e de osteoblastomas atípicos (que apresentam como característica grande celularidade, osteóide azul proeminente e osteoblastos epitelióide—7/30). Como efeito de comparação dos resultados imunohistoquímicos e análise clínica, avaliamos 17 pacientes com osteosarcoma de grau avançado. Os cortes histológicos com bloco de parafina fixado em formalina foram imunocorados para p53 e antígeno nuclear de célula em proliferação. Tumores com coloração positiva para p53 tiveram análise molecular para fragmentos do exon 10.
RESULTADOS:O índice médio de antígeno nuclear de célula em proliferação para osteoblastoma clássico, osteoblastoma atípico e osteosarcoma foram de 33%, 61% e 79%, respectivamente. O subgrupo atípico demonstrou resultados similares aos dos osteosarcomas (p<0,001). Foram detectadas proteína p53 em 4 (13%) osteoblastomas; 3 desses foram osteoblastomas atípicos, sendo que 4 osteosarcomas (23%) também demonstraram p53 positivo. A mutação do DNA nos casos positivos de p53 foi confirmada no exon 10 em dois osteoblastomas atípicos (2/3), um osteoblastoma clássico (1/1) e um osteosarcoma (1/4). A recorrência da doença foi correlacionada com a expressão do p53 (p=0,009), subtipo atípico (p=0,031), osso azul espiculado no resultado da histologia (p=0,018), e índice de marcação pelo antígeno nuclear de célula em proliferação ≥ 40 (p=0,015).
CONCLUSÃO:Esses resultados validam os osteoblastomas atípicos como entidade real, com imunoexpressão das proteínas p53 e antígeno nuclear de célula em proliferação mais perto do osteosarcoma do que do osteoblastoma clássico. O índice de marcação pelo antígeno nuclear de célula em proliferação e o p53 podem ser úteis fatores de prognóstico da recorrência.
Osteoblastoma is an uncommon bone tumor that is considered the benign counterpart of osteosarcoma. Locally aggressive behavior has been noted in some osteoblastomas; however, malignant transformation of the tumors ia rare.1–5 Malignant osteoblastoma or aggressive osteoblastoma,6,7 are terms used to describe some lesions that show atypical cytologic features and may be correlated with recurrence. Osteoblastoma accounts for approximately 1% of all bone tumors.8,9 The most commonly affected sites are the spine, the diaphyses of long bones, pelvis, talus, facial bones, and short bones of the hands and feet. Most of the patients affected by osteoblastoma are in the second or third decades of life.1 Approximately 25% of patients have radiographic and histologic aspects that may suggest malignancy,10 such as large epithelioid osteoblasts, bizarre cells, mitosis, and lace- or sheet-like osteoid production. Several reports6,7,11 have proposed a designation of a borderline osteoblastic tumor, whose radiographic and histologic features vary between the osteoblastoma and osteosarcoma. The term aggressive osteoblastoma was considered preferable because it designates a tumor with local destructive behavior, but without metastatic potential. According to several studies, osteoblastomas may primarily undergo malignant transformation.1,2,4,5 Moreover, cases of osteoblastoma-like osteosarcoma are well-recognized.12,13 Further reports of large series of osteoblastomas have not confirmed the predictive value of the microscopic appearances,1,14 raising the question of whether atypical osteoblastoma really does exist.
The assessment of the proliferative antigen expression yields predictive indicators in soft tissues and bone tumors, with several studies demonstrating the clinical value of anti-PCNA (proliferating cell nuclear antigen) and anti-p53 monoclonal antibodies in these tumors.15–19 Immunohistochemical expression of the p53 protein and the analysis of p53 gene mutations have also been described in 10% to 35% of soft tissue tumors.18 However, only few studies have reported the importance of p53 changes in borderline or low-grade osteoblastic tumors, with controversial results.20–22 Rossner et al23 reported mutation of the p53 gene at exon 5 in a patient with malignant osteoblastoma. Radig et al20 described another mutation at exon 7 in a patient with osteoblastoma-like osteosarcoma. Luca et al24 have previously reported abnormalities in exon 10 of the p53 gene of osteosarcomas, suggesting that exon 10, which is probably involved in the protein oligomerization, is essential for protein function.
The aim of this study was to contribute to understanding this rare bone neoplasm based on morphology, proliferation rate, and p53 mutation analysis. In addition, high-grade osteosarcomas were evaluated to determine discriminating features.
MATERIALS AND METHODSPatientsA retrospective review of 30 patients with osteoblastoma who were treated at the Instituto de Ortopedia e Traumatologia of the São Paulo University Medical School Hospital was performed. The group of patients evaluated consisted of 19 men and 11 women, and the median age was 18.5 years (range, 4-41 years). The most commonly affected sites were the spine (33%) and the long bones of the appendicular skeleton (23%). Typically, a radiolucent and well-circumscribed expansive lesion with calcifications was observed (Figure 1). Seventeen patients with high-grade osteosarcomas were evaluated in order to compare results from immunohistochemical and molecular analyses. In all patients, hematoxylin-eosin–stained sections confirmed the diagnosis. Tumors were classified as atypical osteoblastomas (n = 7) according to the histologic features described by Schajowicz and Lemos7 and Dorfman.6 Schajowicz and Lemos7 proposed the term malignant osteoblastoma for lesions that present with high cellularity, nuclear atypia, and immature tumor bone (spiculated blue bone). Dorfman and Weiss6 proposed the term aggressive osteoblastoma for lesions that present with cellular zones of epithelioid osteoblasts. Radiographic findings and follow-up were documented based on the clinical records.
Immunohistochemistry (IHQ)Deparaffinized formalin-fixed sections were immunostained using monoclonal antibodies to the p53 protein (DO7, 1:2000, DAKO Corporation) and PCNA (PC10, 1:8000, DAKO Corporation). Sections underwent heat-induced epitope retrieval using a pressure-cooker. Antigens were identified using the strepta-avidin-biotin method. For p53, positivity was defined as ≥ 10% of immunostained tumor cells.25 The percentage of tumoral cells positive for PCNA was determined from 500 tumor cells, and results were expressed as PCNA labeling index (PCNA-LI).
Molecular AnalysesPreparing DNA (deoxyribonucleic acid)Genomic DNA was isolated from the archival paraffin-embedded material from the 4 cases of osteoblastoma and the 4 cases of osteosarcoma with positive immunohistochemistry for the p53 protein. Paraffin-block sections from tumor tissues were stained by hematoxylin-eosin to select areas of neoplastic and normal tissues. According to previously published guidelines,26 DNA extraction was performed using paraffin-embedded tissue with the standard phenol chloroform extraction method.
Polymerase chain reaction (PCR)PCR was used to amplify DNA fragments. In all cases, genomic DNA was amplified using a 100-µL PCR mixture of oxynucleotide triphosphate (100 µmol/L), MgCl2 (50 mmol/L), 10 pmol of each of the primers, and Taq polymerase (2.5 U) (Pharmacia Biotech, Uppsala, Sweden). Thermal cycling was performed using a Gene Amp PCR System (Perkin Elmer Cetus, Norwalk, CT). Polymerase chain reaction was done at 94°C for 5 minutes followed by a 30-cycle denaturation at 94°C for 5 minutes, and a 30-cycle denaturation at 94°C for 30 seconds. In all PCR experiments, reactions without DNA were considered negative controls. All PCR products were visualized using 2% ethidium bromide-stained agarose gels. Fragments of exon 10, exon 5, and exon 7 of the p53 with 195 bp were amplified using 2 pairs of oligonucleotide.23 Exon 5: 5’ A T C T G T T C A C T T G T G C C C T G A C T T T C 3’ and 5’ A C C C T G G G C A A C C A G C C C T G T 3’
Exon 10: 5’ G C T G TA T A G G T A C T T G A A G T G C 3’ and 5’ C A T A G G A T T C C A T T C T C A T C 3’
Exon 7: C T C C T A G G T T G G C T C T G 3’ and 5’ G A G G C T G G G G C A C A G C A G G C C A G T G 3’
Direct SequencingPolymerase chain reaction products were prepared for DNA sequencing using exonuclease enzyme (Amersham, Arlington Heights, IL) and shrimp alkaline phosphatase (Amersham) at 37°C for 15 minutes and at 80°C for 15 minutes. Automatic direct DNA sequencing of exon 10 was done using the AB I Prism Dye Terminator Sequencing Kit (Perkin Elmer Cetus for PCR products).
Statistical analysisAnalyses of correlations between immunohistochemical expression of p53 protein, histologic features, and outcomes were carried out using the Fisher exact test. Analyses of the PCNA proliferation index, expression of the p53 protein, and outcomes were done using the Mann-Whitney test. A P value < 0.05 was considered statistically significant. Recurrence was fixed as event of interest, which was evaluated by the Kaplan-Meier test, correlated with p53 immunoexpression, PCNA index ≥ 40 and < 40, and subgroup of osteoblastoma (atypical osteoblastoma x classical osteoblastoma).
RESULTSPathologic findingsHistologically, the major components of the osteoblastoma were osteoblast-like cells, stromal cells, osteoclasts, osteoid tissue deposition, and blood vessels. These cases were classified as classical osteoblastomas (n = 23) (Figure 2). Five patients had destructive infiltration and cortical invasiveness (interruption). In these patients, the most commonly affected sites were the spine, flat bones, and long bones. Atypical osteoblastomas were diagnosed by the following features: epithelioid osteoblasts were identified in 5 patients (Figure 3) and a zone of spiculated blue bone,5 was observed in 6 patients (Figure 4).
Occasionally, osteoblasts showed a focal clustering pattern and hyperchromatic nuclear staining (Figure 5). Cases with features described above were classified as atypical osteoblastomas (n = 7), and the distinction from osteosarcoma was based on the absence of atypical mitotic figures, cellular pleomorphism, neoplastic cartilage, and permeative growth into adjacent bone tissue.13
p53 immunohistochemistry / gene mutationPositive immunoexpression of p53 was identified in 4 cases of osteoblastomas (13.3%), with 3 of these classified as atypical osteoblastoma (Figure 6). Significantly more cases were p53-positive in atypical tumors (P = 0.03). In the osteosarcoma group (n = 17), 4 cases were p53-positive (23.5%). When the 3 groups were compared, immunoexpression of p53 in atypical osteoblastoma was similar to that of osteosarcoma, and both were different from classical osteoblastoma (P < 0.005) (Figure 7). Essentially, expression of the p53 protein was statistically correlated with recurrence (P = 0.009), atypical osteoblastoma classification (P = 0.031), and with the presence of spiculated blue bone (P = 0.018). Mutational changes of the p53 at exon 10 (codon 337) were identified in 3 of 4 patients with osteoblastoma (2 patients with atypical osteoblastoma, and 1 patient with classical osteoblastoma) (Figure 8) and 1of 4 patients with osteosarcoma. Exons 5 and 7 could not be evaluated in the samples because of the limited amount of adequately sized DNA fragments obtained from the archival paraffin embedded tissues, which failed to amplify by PCR.
The atypical osteoblastoma and osteosarcoma groups had higher mean PCNA-LI values than those of classical osteoblastoma (P < 0.001) (Figures 9 and 10). The mean PCNA indexes for classical osteoblastoma, atypical osteoblastoma, and osteosarcoma were 33.7%, 61.7%, and 79.3%, respectively. Results were divided into 2 groups: low PCNA index (< 40%) and high PCNA index (≥ 40%) (30 patients)
The clinical data of 30 patients were available with a 48-month period of follow-up. All patients were treated by total surgical ressection. Five patients (16.6%) had recurrence of the tumor. Three of the patients were in the group with atypical osteoblastoma. Analysis by the Kaplan-Meyer test showed that recurrence was statistically correlated with the atypical osteoblastoma subgroup (P = 0.031), (Figure 11), p53 protein expression (P = 0.009), (Figure 12), presence of spiculated blue bone (P = 0.041), and PCNA labelling index stratified by a 40% cut-off value (P = 0.015) (Figure 13).
The accurate detection and differentiation and treatment of benign and malignant bone tumors can be a serious challenge.27,28 The histologic appearance of osteoblastoma varies widely. At one end of the spectrum are the lesions characterized by trabecular organization of the newly formed osteoid, highly vascular background, and low cellularity. At the other end of this spectrum are the lesions characterized by high cellularity, prominent blue osteoid, mitosis, and epithelioid osteoblasts. These cases may be designated as atypical or aggressive osteoblastomas. In our study, the presence of a lace-like or a sheet-like osteoid deposition was correlated with recurrence (P = 0.041) and aggressive behavior. As with other series, the aggressive pattern of this tumor entity was not correlated with other histological features, such as the evidence of epithelioid osteoblasts and infiltrative histologic growth.1,14
The recurrence rate presented in this series of patients (16.6%) was similar to that of earlier observations.14 The PCNA-LI was correlated with recurrence and the highest indices were associated with atypical groups, as observed in the osteosarcoma group. A PCNA-LI < 40% was rarely found in atypical osteoblastoma (1/7) or osteosarcoma (0/17).
The frequency of p53 immunostaining varied among the different subtypes of osteoblastoma and osteosarcoma, being observed predominantly in osteosarcoma and in atypical osteoblastoma. Interestingly, out of the 2 classical osteoblastomas associated with recurrence, 1 was positive for p53 immunostaining and showed a DNA mutation in exon 10. This is the first study that shows DNA mutation at exon 10 in osteoblastomas. Other studies have also reported p53 mutations and the immunohistochemical protein detection.22,29–31 Immunopositivity for p53 was detected in cases that did not have evidence of p53 gene mutation. This may have occurred for several reasons, including that mutational changes of p53 gene may occur outside the region of the gene analyzed. In this study, Exon 5 and exon 7 were not evaluated in all the samples because the size of the amplified fragments is restricted in archival paraffin-embedded tissues. The quality of DNA extracted from this sort of sample is highly variable, and it depends on decalcification, fixing, and storing of the tissues samples.32
The sample size of the current study was too small to draw definitive conclusions regarding the relationship between clinicopathologic features and p53 changes. However, the results indicating that atypical osteoblastoma, as described by its morphological features, seems to be a real entity. When evaluated by means of proliferation rate and p53 mutations, atypical osteoblastoma exhibits a behavior that is in between classical osteoblastomas and osteossarcomas. The major diagnostic challenge is in differentiating malignant or aggressive osteoblastoma from osteosarcoma on the basis of the pathologic features, since the diagnosis markedly changes treatment in patients.33,34 None of our cases presented metastases. These findings raise the possibility that new genetic events may occur in the group of osteosarcomas that are responsible for its ability to acquire metastatic potential. Our results suggest that the detection of p53 immunopositivity, evidence of a high PCNA labeling index (3 40), and mutations of the p53 gene (exon 10) may have a role in the pathogenesis of osteoblastomas and may reflect the tumoral progression with recurrences, but not exert a sufficient deleterious effect to produce malignant behavior, as is observed in osteosarcoma.
Thus, further investigation, including that regarding functional analysis of mutations, is essential to demonstrate the role of genetic changes in the clinical and histopathologic features of osteoblastoma.
ACKNOWLEDGMENTSWe wish to thank Dr Nany Renzo Barbosa de Oliveira for providing archival tumor material and for her expert help in interpreting the diagnosis, Rita de Cassia A. M. G. Pinto for supporting technical assistance in immunohistochemistry, Dr. Katia R. M. Leite, for suggestions in molecular biology procedures, Carlos Fernando Saito for organizing the tables and Natalina F. A. Santos for her assistance in writing this manuscript.