In breast cancer (BC) patients, the frequency of germline BRCA mutations (gBRCA) may vary according to the ethnic background, age, and family history of cancer. Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA) is the second most common somatic mutated gene in BC; however, the association of mutations in both genes with cancer has not been thoroughly investigated. Thus, our aims were to investigate gBRCA mutation frequency in a cohort of postmenopausal Brazilian BC patients and the association of gBRCA1/BRCA2 and PIK3CA somatic mutations.
METHODS:Forty-nine postmenopausal (>55 years) and forty-one young (≤35 years) BC patients were included in this study. The postmenopausal group included patients who reported a positive family history of cancer. For these patients, gBRCA1/BRCA2 were sequenced using next-generation sequencing (NGS) or Sanger sequencing. Data for gBRCA in young patients were already available from a previous study. DNA from formalin-fixed, paraffin-embedded (FFPE) tumors was obtained from 27 postmenopausal and 41 young patients for analyzing exons 9 and 20 of PIK3CA. The association between gBRCA1/BRCA2 and somatic mutations in PIK3CA was investigated.
RESULTS:The overall frequency of gBRCA1/BRCA2 among the 49 postmenopausal patients was 10.2%. The frequencies of somatic mutations in PIK3CA in the postmenopausal and young patients were 37% and 17%, respectively (ns). The most common PIK3CA mutation was found to be E454A. Nonsense and frameshift mutations, which may counteract the oncogenic potential of PIK3CA were also detected. Regardless of age, 25% of BRCA1/BRCA2 mutation carriers and non-carriers , each, had PIK3CA somatic mutations.
CONCLUSIONS:Data obtained indicate that BRCA1/BRCA2 gene testing may be considered for postmenopausal patients with BC who have a family history of cancer. Although some of them are not considered pathogenic, somatic variants of PIK3CA are frequently observed in BC patients, especially in postmenopausal patients.
Breast cancer affects women of all ages; however, the incidence of breast cancer increases with age, and the peak incidence occurs between 45-64 years (1). In addition, breast cancer is the most prevalent cancer in women aged 30-39 years (2). The main risk factors for breast cancer are a) age, b) positive family history of breast and ovarian cancer, and c) hormone exposure (3).
A positive family history is observed in approximately 10-20% of the breast cancer patients, but mutations in predisposing genes have been identified in <30% of these cases (4). BRCA1/BRCA2—both related to the homologous repair of DNA double-strand breaks—are the major breast/ovarian cancer susceptibility genes. Generally, women who harbor BRCA1/BRCA2 mutations are more frequently diagnosed with breast cancer at an early age (≤40 years) or with ovarian cancer at any age. In addition, women who develop breast cancer at an older age and report a strong family history of breast/ovarian cancer mainly in close relatives—first, second, or third degree—may also be BRCA1/BRCA2 mutation carriers (5). However, the majority of breast cancer cases are sporadic, i.e., not related to genetic syndromes. In this case, somatic mutations accumulate over an individual's lifetime, similar to an ‘evolutionary’ process, a phenomenon that makes age itself a risk factor for cancer (6). In this process, some cells acquire mutations that are advantageous from a tumoral perspective, which allows aberrant proliferation, invasion, and metastasis.
In breast cancer, somatic mutations in the PIK3CA gene are the most frequent, just after TP53 (7). The PIK3CA gene encodes the p110 catalytic subunit of a heterodimeric lipid kinase called PI3K that is activated in response to various extracellular signals that are transduced through receptor tyrosine kinases. After activation, PI3K phosphorylates phosphatidylinositol-4,5-bisphosphate (PI-4,5-P2), generating phosphatidylinositol-3,4,5-trisphosphate (PIP3), which functions as a second messenger and recruits proteins that harbor pleckstrin homology (PH) domains (e.g., AKT) (8). Mutations in the helical or kinase domain of PIK3CA resulted in the activation of the p110a kinase, with the subsequent downstream activation of mediators that culminates in cell proliferation, angiogenesis, and promotion of metastasis (9,10).
In breast cancer, an association between somatic mutations in PIK3CA and the positive expression of the estrogen receptor (ER) has been reported (11–14). However, the association between the frequency of somatic mutations in PIK3CA and age is unclear (15,16). Moreover, it seems likely that the frequency of somatic mutations in PIK3CA increases in ER-positive tumors in aging patients (7).
Thus, BRCA1 and BRCA2 are the most common germline mutated genes, while PIK3CA is the second most common somatic mutated gene in breast cancer patients; however, subtle frequency differences may be related to the age of onset of the disease. Carcinogenic mechanisms elicited by BRCA1/BRCA2 loss of function and PIK3CA gain of function may be targeted for therapy. There is evidence that combination therapies targeting tumors harboring BRCA mutations—such as PARP inhibitors—with PI3K pathway inhibition therapies may exhibit synergy in vivo for the treatment of endogenous BRCA1-related breast cancer mouse model (17). However, it has been previously reported that the frequency of PIK3CA mutations may be different in breast cancer patients based on the presence of germline mutations in BRCA1/BRCA2 (in both women and men) (18,19). Thus, our aim was to investigate the frequency of BRCA mutations in a cohort of postmenopausal Brazilian breast cancer patients, for whom scarce information is available. The secondary exploratory aim of this study was to evaluate the association of germline BRCA1/BRCA2 mutations with somatic PIK3CA mutations in a cohort of young and postmenopausal patients with breast cancer.
METHODSPatientsPatients were recruited at the Instituto do Câncer do Estado de São Paulo (ICESP), the cancer treatment branch of Hospital das Clínicas da Faculdade de Medicina da Universidade de São Paulo, the largest public hospital complex in Latin America, São Paulo, Brazil. This study was approved by the Institutional Ethics Committee (Comitê de Ética da Faculdade de Medicina da Universidade de São Paulo; protocol 397/11). All patients signed informed consent forms.
The inclusion criteria were 1) histopathological diagnosis of invasive breast carcinoma in patients aged <36 years or >54 years; 2) patients aged 55 years or older with at least one relative having first, second, or third degrees and diagnosed with breast, ovarian pancreatic, or prostate cancer; 3) triple-negative tumor and age ≤60 years. The expression of hormone receptor was classified as positive if at least 1% of the malignant cells were stained with antibodies against estrogen or progesterone receptor; HER2 positivity was defined as immunohistochemistry scores of 3(+) or 2(+), the latter, associated with fluorescence in situ hybridization (FISH)-amplification. HER2 immunohistochemistry and FISH were scored according to the ASCO/CAP guidelines (20). The Ki67 expression cut-off was set at >14% for a high proliferation index. The molecular subtypes were classified using previously established criteria (21).
Personal and familial cancer histories were collected through a structured questionnaire. Patients were also asked about their ancestry to obtain information about the country or continent where their parents and grandparents (at least) were born. A pedigree that reached up to third-degree relatives was designed. Clinical and pathological data were retrieved from hospital files.
In a previous study, 79 very young breast cancer patients (≤35 years) were evaluated for the presence of germline mutations in BRCA1 and BRCA2, among whom, four harbored BRCA1 mutations (c.66_67insA; c.211A>G; c.3331_3334delCAAG; c.5263_5264insC) and nine harbored BRCA2 mutations (c.483T>A; c.1138_1138delA; c.2808_2811delACAA (n=2); c.3956_3959delATGA; c.6656C>G; c.6990_6994delTACCT; c.9154C>T; c.9382C>T) (22). For detecting PIK3CA mutations, tumor samples were available for 41 patients (among the 79 patients) and were included in the present analysis. Clinical data and tumor subtypes based on ER, PR, HER2, and Ki67 expression levels (as described above) are summarized in Table 4 (22). Six of these forty-one patients harbored BRCA1 or BRCA2 mutations.
Clinical and pathological features of breast cancer patients according to their age.
| Features | Postmenopausal | Young | |
|---|---|---|---|
| n=27 | n=41 | p | |
| Age at diagnosis, median (range), years | 61 (55-74) | 32 (23-35) | |
| Tumor Subtype | |||
| Luminal A | 8 (8) | 2 (4.9) | 0.04 |
| Luminal B | 14 (51.9) | 19 (46.3) | |
| Luminal | 2 (7.4) | 4 (9.8) | |
| HER2+ | 1 (3.7) | 5 (12.2) | |
| Triple Negative | 2 (7.4) | 10 (24.4) | |
| Not Determined | 0 | 1 (2.4) | |
| Clinical Stage, n (%) | |||
| I/II | 19 (73.1) | 23 (65.7) | 0.539 |
| III/IV | 7 (26.9) | 12 (34.3) | |
| BRCA germline status | |||
| BRCA1/BRCA2 mut | 2 (7.4) | 6 (14.6) | 0.365 |
| BRCA1/BRCA2 wt | 25 (92.6) | 35 (85.4) | |
| PIK3CA somatic status | |||
| PIK3CA path mut | 10 (37) | 7(17.1) | * |
| PIK3CA wt | 17 (63) | 34 (82.9) | |
| Luminal Tumors vs PIK3CA somatic status | |||
| Luminal PIK3CA mut | 8 (33.3%) | 5 (20%) | 0.291 |
| Luminal PIK3CA wt | 16 (66.7%) | 20 (80%) | |
Tumor Subtype based on ER, PR, HER2 and Ki67 expression, as described in methods. Missing data were not computed. Pearson's chi-Square. *not tested owing to the small sample size.
Genomic DNA from peripheral blood samples was extracted using the Illustra Blood Genomic Prep Mini Spin Kit (GE Healthcare Bio-Sciences, Pittsburgh, PA, USA), and from cancer cell-enriched areas from the formalin-fixed, paraffin-embedded (FFPE) tumor samples using the QIAamp® DNA FFPE Tissue (Qiagen, Valencia, CA, USA), as per the manufacturer’s protocol.
DNA concentration and purity were determined using a NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific, Massachusetts, USA), and the absorbance260/280 ratio varied from 1.42 to 2.2. DNA concentration from samples analyzed using next-generation sequencing (NGS) was also evaluated using a Qubit® dsDNA BR Assay kit on a Qubit® 3.0 Fluorometer (Invitrogen, Carlsbad, California, USA).
Analysis of Germline Mutations in BRCA1/BRCA2The entire coding regions of BRCA1 and BRCA2, including exon-intron boundaries, were sequenced by NGS using the Ion Torrent Personal Genome Machine (PGM) platform (n=38) or by Sanger sequencing (n=11), to determine the presence of germline mutations.
Next-Generation SequencingBRCA1 and BRCA2 were sequenced using the Ion AmpliSeq™ BRCA1 and BRCA2 Panel (Life Technologies, Carlsbad, CA, USA) consisting of three primer pools, covering the target regions in 167 amplicons that target the entire coding region, including 10-20 bp of non-coding sequences, flanking the 5‘ and 3‘ ends of each exon, for both genes. Libraries containing the PCR product were sequenced on a 314 v2 Ion Chip, which allows the simultaneous analysis of 12 samples per chip on a PGM sequencer (Ion Torrent™), and the Ion PGM Sequencing 200 Kit version 2 (Life Technologies, Carlsbad, CA, USA). Data analysis was performed using the Ion Reporter™ Server System (Thermo Fisher Scientific, Massachusetts, USA). Sequence data were also visually evaluated using the Integrative Genomics Viewer (IGV). Amplicons with coverage less than 30x, pathogenic variants, and new variants were confirmed by PCR followed by conventional bidirectional Sanger sequencing. Full details of the methods are provided in the Appendix.
PCR and Sanger SequencingAll coding regions, including the intron-exon boundaries of BRCA1 (NM_7294.3) and BRCA2 (NM_000059.3) were amplified by PCR. Primers and conditions are described in the Appendix. The amplicons were purified (Illustra™ ExoStar™ 1-Step-GE Healthcare Bio-Sciences, Pittsburgh, PA, USA) and were sequenced using the BigDyeTM Terminator v3.1 Cycle Sequencing kit (Applied BiosystemsTM, Foster City, California, USA), as described previously (22). Following purification, samples were analyzed on a 3500 Genetic Analyzer or ABI 3730 DNA Analyzer (Applied Biosystems™, Foster City, California, USA) in both forward and reverse directions (Appendix). The results were analyzed using Mutation Surveyor DNA Variant Analysis Software (v3.30, SoftGenetics LLC). All pathogenic mutations were confirmed using Sanger sequencing.
Analysis of Copy Number Variation in BRCA1 and BRCA2For the analysis of large deletions and duplications—that would have provided comprehensive information regarding germline mutations—patient DNA was subjected to BRCA1 and BRCA2 multiplex ligation-dependent probe amplification (MLPA) analysis (BRCA1: SALSA® MLPA® P002 and P087 Probemix; BRCA2: SALSA® MLPA® P045 BRCA2/CHEK2 Probemix; MRC-Holland, Amsterdam, The Netherlands), as per the manufacturer’s protocols (Appendix), as described previously (22,23).
Mutation Nomenclature and ClassificationBRCA1 and BRCA2 variants were named according to the Human Genome Variation Society (HGVS) nomenclature (24) and were searched in publicly accessible databases, i.e., BRCA Share™, BRCA Exchange, BRCA Mutation Database, and ClinVar. The search was performed in 2020 (between January and June). In silico analyzes were performed using the following prediction tools: Polymorphism Phenotyping (PolyPhen; v2.2.2), Sorting Intolerant From Tolerant (SIFT; v1.0.3), Align-GVGD, Protein Variation Effect Analyzer (Provean; v1.1), and Human Splicing Finder to analyze variants of unknown clinical significance. Minor allele frequency (MAF) was checked on the 1000 Genomes Project database, Exome Aggregation Consortium (ExAC), Global MAF dbSNP, Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP), Genome Aggregation Database (gnomAD), Trans-Omics for Precision Medicine (TOPMed), and Brazilian genomic variants (ABraOM). More details are provided in the Appendix.
The variants were then classified as pathogenic, likely pathogenic, benign, likely benign, and variant of uncertain significance (VUS) based on the recommendations of the American College of Medical Genetics and Genomics (25). VUS for BRCA was also checked for co-occurrence with known pathogenic mutations in the same patient. For some variants, we considered that consensus information in ≥2 databases was strong enough to classify them as benign or VUS.
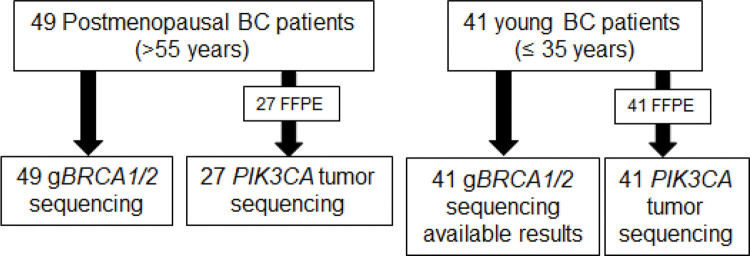
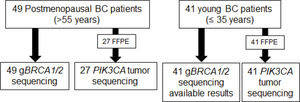
Analysis of Somatic Mutations in PIK3CAAmong the 49 postmenopausal patients, 27 FFPE tumor samples were available for analysis. Tumor samples from another 22 patients were not available because they had been operated on at another service. Tumor samples from all 41 young patients were used for further analysis (Figure 1).
PIK3CA (NM_006218.2) exons 9 (helical domain) and 20 (kinase domain), which are the regions with the highest mutation frequency (26), were amplified by PCR and were analyzed by Sanger sequencing. Primer sets were designed using software Primer3 (http://bioinfo.ut.ee/primer3/). To avoid non-specific product formation, BLAST (http://www.ncbi.nlm.nih.gov/blast) and BLAT (https://genome.ucsc.edu/cgi-bin/hgBlat) were performed. Primers and conditions are described in the Appendix.
Statistical Analysis and Sample Size CalculationTo detect the frequency of germline BRCA mutations in postmenopausal breast cancer patients (varying from 2% to 17%), a sample size of 50 was estimated (27,28). For analyzing the frequency of PIK3CA mutations in young and postmenopausal patients; this was a convenient sample size, because only 55% of tumor samples were available for the latter. Assuming that the frequency of PIK3CA mutations in young and postmenopausal patients was 7% and 35%, respectively (7) and the correlation of two postmenopausal patients for every three young patients, the estimated sample size to detect a difference with 0.05 one-sided significance level and 80% power would be 31 young and 21 postmenopausal patients.
Pearson's chi-square test was used to evaluate the association between variables, and a two-sided significance level of 0.05 was considered.
RESULTSPatientsForty-nine elderly women aged ≥55 years who were diagnosed with invasive ductal breast carcinoma were included between May 2014 and May 2015 and evaluated for the presence of germline mutations in BRCA. FFPE tumor samples of 27 patients were analyzed for the presence of somatic mutations in PIK3CA. The median ages at the time of diagnosis and enrollment in the study were 61 years (55-80 years) and 64 years (56-87 years), respectively. The majority of the patients had Nottingham histological grade II tumors (63.3%) and clinical stage I/II tumors (67.4%). With respect to the tumor subtype, most tumors were luminal B (44.9%) or luminal A (22.4%), followed by HER2+ and triple-negative tumors (10.2% each) (Table 1; Additional Table 1). Most patients (95.9%)—except for two patients (one with a triple-negative tumor and age ≤60 years)—reported a positive family history of breast, ovarian, pancreatic, or prostate cancers. A large proportion of the patients (69.4%) reported at least one affected first-degree family member with breast and/or ovarian cancer. Most women were born in the Southeast (67.3%)—followed by the Northeast (18.4%)—regions of Brazil. With respect to ancestry, 28.6% of the patients reported Brazilian and European ancestries, 26.5% reported only Brazilian ancestry, and 18.4% and 8.4% reported European-only or Asian ancestry, respectively (Table 1).
Clinical and pathological features of breast cancer patients according to deleterious BRCA1 and BRCA2 mutations.
| Features | BRCA1/BRCA2 mut | BRCA1/BRCA2 wt | |
|---|---|---|---|
| n=49 | n=5 | n=44 | |
| Age at diagnosis, median (range), years | 61 (55-80) | 58 (56-80) | 62 (55-80) |
| Age at enrollment, median (range), years | 64 (56-87) | 60 (58-82) | 64.5 (56-87) |
| Histological grade, n (%) | |||
| I | 10 | 0 | 10 (100) |
| II | 31 | 2 (6.5) | 29 (93.5) |
| III | 7 | 3 (42.8) | 4 (57.8) |
| Missing | 1 | 0 | 1 (100) |
| Clinical Stage, n (%) | |||
| I | 14 | 0 | 14 (100) |
| II | 19 | 1 (5.3) | 18 (94.7) |
| III | 10 | 2 (20) | 8 (80) |
| Missing | 6 | 2 (33.5) | 4 (66.5) |
| Molecular Subtype | |||
| Luminal A | 11 | 0 | 11 (100) |
| Luminal B | 22 | 3 (13.7) | 19 (86.4) |
| Luminal | 6 | 0 | 6 (100) |
| HER2+ | 5 | 0 | 5 (100) |
| Triple Negative | 5 | 2 (40) | 3 (60) |
| Affected relatives, n (%) | |||
| First Degree | 34 | 4 (11.8) | 30 (82.2) |
| Second Degree | 9 | 0 | 9 (100) |
| Third Degree | 4 | 1 (25) | 3 (75) |
| Negative | 2 | 0 | 2 (100) |
| Ancestry until second degree, n (%) | |||
| Brazilian only | 13 | 2 (15.4) | 11 (84.6) |
| European only | 9 | 0 | 9 (100) |
| Asian only | 5 | 1 (20) | 4 (80) |
| Brazilian and European | 14 | 1 (7.2) | 13 (92.8) |
| Brazilian and Indigenous | 1 | 0 | 1 (100) |
| Brazilian and Australian | 1 | 1 (100) | 0 |
| Brazilian and South American | 1 | 0 | 1 (100) |
| Brazilian and European and Australian | 1 | 0 | 1 (100) |
| Indigenous and European | 1 | 0 | 1 (100) |
| European and Unknown | 1 | 0 | 1 (100) |
| Indigenous and Unknown | 1 | 0 | 1 (100) |
| Unknown | 1 | 0 | 1 (100) |
| Region of origin, n (%) | |||
| Southeast | 33 | 2 (6) | 31 (94) |
| Northeast | 9 | 2 (22.2) | 7 (77.8) |
| South | 3 | 0 | 3 (100) |
| Abroad | 4 | 1 (25) | 3 (75) |
Clinical and pathological characteristics of breast cancer patients, BRCA sequencing, and the multiplex ligation-dependent probe amplification (MLPA) results.
| ID | Age Years | HT | HG | ER (%) | PR (%) | HER2 | Ki67 (%) | Molecular Subtype | CS | FH | BRCA | MLPA |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 71 | IDC | 2 | 100 | 80 | Neg. | 25 | Luminal B | III | Yes | wt | wt |
| 2 | 80 | IDC | 2 | 80 | 80 | Neg. | 30 | Luminal B | ND | Yes | BRCA2 | wt |
| 3 | 66 | IDC | 1 | 95 | 80 | Neg. | 15 | Luminal B | II | Yes | wt | ND |
| 4 | 61 | IDC | 2 | 95 | Neg. | Neg. | 20 | Luminal B | II | Yes | wt | wt |
| 5 | 61 | IDC | 2 | 100 | 100 | Neg. | 12 | Luminal A | II | Yes | wt | wt |
| 6 | 74 | IDC | 1 | 100 | 100 | Neg. | 10 | Luminal A | I | Yes | wt | wt |
| 7 | 66 | IDC | 2 | 60 | 66 | Neg. | 30 | Luminal B | I | Yes | wt | wt |
| 8 | 61 | IDC | 2 | Pos. | Pos. | Neg. | 30 | Luminal B | III | Yes | wt | wt |
| 9 | 73 | IDC | 2 | 100 | 100 | Neg. | 10 | Luminal A | I | Yes | wt | wt |
| 10 | 57 | IDC | ND | 90 | 70 | Neg. | 10 | Luminal A | I | Yes | wt | wt |
| 11 | 73 | IDC | 2 | 95 | Neg. | Neg. | 15 | Luminal B | II | Yes | wt | wt |
| 12 | 73 | IDC | 3 | 100 | 5 | Neg. | ND | Luminal | I | Yes | wt | wt |
| 13 | 59 | IDC | 1 | Neg. | Neg. | Pos. | 18 | HER 2 | III | Yes | wt | wt |
| 14 | 62 | IDC | 2 | 90 | 70 | Neg. | ND | Luminal | II | Yes | wt | wt |
| 16 | 60 | IDC | 1 | 90 | 100 | Neg. | 8 | Luminal A | II | Yes | wt | wt |
| 17 | 56 | IDC | 3 | Pos. | Pos. | Neg. | 80 | Luminal B | III | Yes | BRCA1 | wt |
| 18 | 56 | IDC | 3 | Neg. | Neg. | Neg. | 30 | TN | I | Yes | wt | wt |
| 19 | 63 | IDC | 2 | 10 | Neg. | Neg. | 20 | Luminal B | I | Yes | wt | wt |
| 20 | 65 | IDC | 2 | 50 | Neg. | Neg. | 30 | Luminal B | II | Yes | wt | wt |
| 21 | 67 | IDC | 1 | 100 | 100 | Neg. | 30 | Luminal B | II | Yes | wt | wt |
| 22 | 56 | IDC | 2 | 66 | 1 | Neg. | 30 | Luminal B | II | Yes | wt | wt |
| 23 | 62 | IDC | 2 | 95 | 1 | Neg. | 18 | Luminal B | I | Yes | wt | wt |
| 24 | 76 | IDC | 3 | Neg. | Neg. | Pos. | 40 | HER2 | III | Yes | wt | wt |
| 25 | 60 | IDC | 2 | Neg. | Neg. | Neg. | 65 | TN | ND | Yes | wt | wt |
| 26 | 60 | IDC | 2 | Pos. | Neg. | Neg. | 30 | Luminal B | II | Yes | wt | wt |
| 27 | 56 | IDC | 2 | Pos. | Pos. | Neg. | 30 | Luminal B | ND | Yes | wt | wt |
| 28 | 63 | IDC | 2 | 66 | 66 | Neg. | 30 | Luminal B | ND | Yes | wt | wt |
| 29 | 58 | IDC | 2 | Neg. | Neg. | Neg. | 33 | TN | ND | Yes | BRCA1 | wt |
| 30 | 62 | IDC | 1 | 100 | Neg. | Neg. | 5 | Luminal A | III | Yes | wt | wt |
| 31 | 60 | IDC | 2 | 66 | 66 | Neg. | 30 | Luminal B | ND | Yes | wt | wt |
| 32 | 56 | IDC | 1 | 90 | 70 | Neg. | 10 | Luminal A | I | No | wt | wt |
| 33 | 55 | IDC | 1 | Neg. | Neg. | Neg. | ND | TN | I | No | wt | wt |
| 34 | 61 | IDC | 2 | 95 | 15 | Neg. | 20 | Luminal B | II | Yes | wt | wt |
| 35 | 68 | IDC | 2 | 66 | 66 | Neg. | 10 | Luminal A | II | Yes | wt | wt |
| 36 | 63 | IDC | 1 | 90 | 80 | Neg. | 10 | Luminal A | I | Yes | wt | wt |
| 37 | 62 | IDC | 2 | 95 | 0,1 | Pos. | 40 | Luminal B | I | Yes | wt | wt |
| 38 | 59 | IDC | 2 | 40 | 75 | Neg. | 20 | Luminal B | I | Yes | wt | wt |
| 39 | 63 | IDC | 2 | 100 | 100 | Neg. | 13 | Luminal A | II | Yes | wt | wt |
| 40 | 63 | IDC | 2 | 100 | 30 | Pos. | 20 | Luminal B | III | Yes | wt | wt |
| 41 | 62 | IDC | 2 | Pos. | Pos. | Pos. | ND | Luminal | II | Yes | wt | wt |
| 42 | 77 | IDC | 3 | Neg. | Neg. | Pos. | 70 | HER2 | III | Yes | wt | wt |
| 43 | 65 | IDC | 2 | >50 | >50 | Neg. | 5-30 | Luminal | II | Yes | wt | wt |
| 44 | 56 | IDC | 3 | 90 | Neg. | Neg. | 30-40 | Luminal B | II | Yes | BRCA2 | wt |
| 45 | 64 | IDC | 2 | Pos. | Pos. | Neg. | ND | Luminal | II | Yes | wt | wt |
| 46 | 55 | IDC | 2 | Neg. | Neg. | Pos. | 10 | HER2 | III | Yes | wt | wt |
| 47 | 58 | IDC | 3 | Neg. | Neg. | Neg. | 70 | TN | III | Yes | wt | BRCA1 |
| 48 | 75 | IDC | 1 | >66 | >66 | Neg. | <15 | Luminal A | I | Yes | wt | ND |
| 49 | 79 | IDC | 2 | Neg. | Neg. | Pos. | 40 | HER2 | II | Yes | wt | wt |
| 50 | 80 | IDC | 2 | Pos. | Pos. | Neg. | ND | Luminal | II | Yes | wt | wt |
ID: Patient identification; HT: Histological type; HG: Histological grade; ER: Estrogen receptor; PR: Progesterone receptor; CS: Clinical stage; FH: Family history for breast and/or ovarian cancer; ND: Not determined; wt: Wild type; MLPA: Multiplex ligation-dependent probe amplification.
Another 41 young patients, aged ≤35 years, had their tumor samples evaluated for the presence of somatic mutations in PIK3CA. This is a subgroup of patients whose clinical data, as well as germline BRCA1 and BRCA2 sequencing results had already been reported in a previous study (22). The cohort of patients now reported comprehends those young patients who had FFPE tumor samples available for PIK3CA analysis. The median age at the time of diagnosis was 32 years (range, 23-35 years). Most patients presented tumors with histological grade II (43.9%) or III (48.8%), and disease clinical stage I/II (65.7%). Luminal B (46.3%) was the most frequent tumor subtype, followed by triple-negative (24.4%) and HER2 (+) (12.2%) tumors (Table 4). Among these patients, 14.6% and 12.2% reported first-or second-degree relatives diagnosed with breast and/or ovarian cancer, respectively, while 39% reported a negative family history of breast and/or ovarian cancer, and 24.4% were not able to describe their family history. Six out of the forty-one patients harbored pathogenic mutations (14.6%) in BRCA1 or BRCA2, as previously reported (22).
Germline Mutations in BRCA1 and BRCA2 in Postmenopausal PatientsAmong 49 postmenopausal unrelated women, 5 (10.2%) were identified to harbor mutations of clinical significance, 3 in BRCA1 and 2 in BRCA2 (Table 2; Additional Tables 2-3). All five BRCA mutations were identified among 47 patients who reported a positive family history of breast, ovarian, prostate, and pancreatic cancers in close relatives (10.6%), including four mutations detected among 34 patients reporting first-degree relatives affected by these types of cancer (11.76%) (Table 1).
BRCA1 and BRCA2 mutations in breast cancer patients: Clinical aspects and molecular description.
| ID | HGVS cDNA | HGVS protein | Type | BrCa Age | OvCa Age | Tumor Subtype | HG | CS | Ancestry | FH |
|---|---|---|---|---|---|---|---|---|---|---|
| BRCA1 | ||||||||||
| 29 | c.5074+2T>C | − | SS | 58 | − | TN | 2 | ND | BRZ | Pos |
| 17 | c.5123C>A | p.Ala1708Glu | M | 56 | − | Lum B | 3 | III | BRZ/AUS | Pos |
| 47 | Exon 1-19 deleted | − | LGR | 58 | − | TN | 3 | III | BRZ/EUR | Pos |
| BRCA2 | ||||||||||
| 44 | c.2T>G | p.Met1Arg | M | 56 | − | Lum B | 3 | II | BRZ | Pos |
| 2 | c.5645C>A | p.Ser1882Ter | NS | 80 | >70 | Lum B | 2 | ND | Asian | Pos |
ID: Patient identification; SS: Splice site; M: Missense; LGR: Large genomic rearrangement; NS: Nonsense; Lum: Luminal; HG: Histological grade; CS: Clinical stage; AUS: Australian; FH: Family history of breast, ovarian, pancreatic or prostate cancer; Pos: Positive.
BRCA1 variants.
| Exon | HGVS Nucleotide | HGVS Protein | Protein | Other names | Type | Localization (GRCh37) | NCBI 1000 Genomes Browser | Global MAF dbSNP | Allele Frequency ExAC | Global MAF 1000 genomes | ESP | gnomAD | TOPMed | ABraOM | SIFT | PolyPhen | Provean | Align-GVGD (Pufferfish) | Human Splicing Finder | BRCA Exchange | BRCA Mutation Database | BRCA Share™ | ClinVar | Interpretation | n |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | c.-19-115T>C | − | − | IVS1-115T>C | 5'UTR | 17: 41276247 | rs3765640 | 0.35363 (G) | − | 0,35363 | − | 0,31688 | 0,30248 | 0.304260 | − | − | − | − | Mutant type not implemented in HSF yet | Benign/Little Clinical Significance | ND | ND | Benign | Benign | 9 |
| 2 | c.81-14C>T | − | − | IVS2-14C>T | IVS | 17: 41267810 | rs80358006 | − | − | − | 0.00069 | − | 0,00052 | 0.001642 | − | − | − | − | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign/Likely Benign | Benign/Likely Benign | 1 |
| 3 | c.134+111C>T | − | − | IVS3+111C>T | IVS | 17: 41267632 | rs8176100 | 0.00379 (A) | − | 0,00379 | − | 0,00227 | 0.00128 | − | − | − | − | − | Creation of an intronic ESE site. Probably no impact on splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 1 |
| 6 | c.301+43A>G | − | − | IVS6+43A>G | IVS | 17: 41256841 | − | − | − | − | − | − | − | − | − | − | − | − | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | ND | ND | ND | ND | Uncertain Significance | 1 |
| 7 | c.441+36_441+49delCTTTTCTTTTTTTT | − | − | IVS7+36del14 | IVS | 17: 41256090_41256103 | rs373413425 | − | − | − | − | − | − | 0.295230 | − | − | − | − | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Not Yet Reviewed | ND | 1-Neutral | Benign | Benign | 23 |
| 7 | c.441+36C>T | − | − | IVS7+36C>T | IVS | 17: 41256103 | rs45569832 | − | − | − | − | 0,00009 | − | − | − | − | − | − | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Not Yet Reviewed | ND | 3-UV | Uncertain significance? | Uncertain Significance | 2 |
| 7 | c.441+41C>T | − | − | IVS7+41C>T | IVS | 17: 41256098 | rs45489593 | − | 0,00024 | − | − | 0,00104 | − | − | − | − | − | − | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Not Yet Reviewed | ND | 1-Neutral | Uncertain significance? | Uncertain Significance | 1 |
| 7 | c.442-34C>T | − | − | IVS7-34C>T | IVS | 17: 41251931 | rs799923 | 0.09864 (A) | 0,17379 | 0,09864 | 0,17569 | 0,17303 | 0,14802 | 0.200328 | − | − | − | − | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Not Yet Reviewed | ND | 1-Neutral | Benign | Benign | 15 |
| 9 | c.548-58delT | − | − | c.IVS8-58delT | IVS | 17: 41249364 | rs8176144 | 0.33486 (AAAAAA) | − | − | 0,27833 | 0.3005 | 0,28382 | − | − | − | − | − | Alteration of an intronic ESS site. Probably no impact on splicing. | ND | ND | 1-Neutral | Benign | Benign | 2 |
| 9 | c.591C>T | p.Cys197= | C197= | 710C>T | Syn | 17: 41249263 | rs1799965 | 0.00040 (A) | 0,00147 | 0.00040 | 0,00123 | 0,00178 | 0,00076 | − | − | − | Neutral | − | Activation of an exonic cryptic donor site. Creation of an exonic ESS site. Potential alteration of splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 1 |
| 11 | c.1067A>G | p.Gln356Arg | Q356R | 1186A>G | M | 17: 41246481 | rs1799950 | 0.02177 (C) | 0,04407 | 0,02177 | 0,0459 | 0,05196 | 0,04129 | 0.049261 | Deleterious (0.01) | Probably Damaging (0.988) | Deleterious | Class C0 | Creation of an exonic ESS site. Alteration of an exonic ESE site. Potential alteration of splicing. | Benign/Little Clinical Significance | 1-Not pathogenic or of no clinical significance | 1-Neutral | Benign | Benign | 6 |
| 11 | c.1971A>G | p.Gln657= | Q657= | 2090 A>G | Syn | 17: 41245577 | rs28897679 | 0.00639 (C) | 0,00217 | 0,00639 | 0,00869 | 0,00605 | 0,00741 | 0.005747 | − | − | Neutral | − | Creation of an exonic ESS site. Alteration of an exonic ESE site. Potential alteration of splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 1 |
| 11 | c.2077G>A | p.Asp693Asn | D693N | 2196G>A | M | 17: 41245471 | rs4986850 | 0.03355 (T) | 0.05681 | 0,03355 | 0,05429 | 0.05451 | 0.05336 | 0.056650 | Tolerated (0.08) | Benign (0.01) | Neutral | Class C0 | Alteration of an exonic ESE site. Potential alteration of splicing. | Benign/Little Clinical Significance | 1-Not pathogenic or of no clinical significance | 1-Neutral | Benign | Benign | 9 |
| 11 | c.2082C>T | p.Ser694= | S694= | 2201C>T | Syn | 17: 41245466 | rs1799949 | 0.33646 (A) | 0,34827 | 0,33646 | 0,29568 | 0,31633 | 0,30145 | 0.302956 | − | − | Neutral | − | Activation of an exonic cryptic donor site. Alteration of an exonic ESE site. Potential alteration of splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 22 |
| 11 | c.2311T>C | p.Leu771= | L771L | 2430T>C | Syn | 17: 41245237 | rs16940 | 0.33526 (G) | 0,34196 | 0,33526 | 0,27764 | 0,30018 | 0,28384 | 0.282430 | − | − | Neutral | − | Creation of an exonic ESS site. Potential alteration of splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 22 |
| 11 | c.2596C>T | p.Arg866Cys | R866C | 2715C>T | M | 17: 41244952 | rs41286300 | − | 0,0001 | − | − | 0,00016 | 0,00017 | − | Deleterious (0) | Probably Damaging (1) | Deleterious | Class C65 | Creation of an exonic ESS site. Potential alteration of splicing. | Benign/Little Clinical Significance | 1-Not pathogenic or of no clinical significance | 1-Neutral | Benign | Benign | 1 |
| 11 | c.2612C>T | p.Pro871Leu | P871L | 2731C>T | M | 17: 41244936 | rs799917 | 0.45607 (G) | 0,41005 | 0,54393 | 0,49316 | − | 0,4893 | 0.450739 | Tolerated (1) | Benign (0) | Neutral | Class C0 | Creation of an exonic ESS site. Alteration of an exonic ESE site. Potential alteration of splicing. | Benign/Little Clinical Significance | 1-Not pathogenic or of no clinical significance | 1-Neutral | Benign | Benign | 26 |
| 11 | c.3113A>G | p.Glu1038Gly | E1038G | 3232A>G | M | 17: 41244435 | rs16941 | 0.33566 (C) | 0,34287 | 0,33566 | 0,27903 | 0,30081 | 0,28456 | 0.282430 | Tolerated (0.16) | Possibly Damaging (0.606) | Deleterious | Class C0 | Creation of an exonic ESS site. Alteration of an exonic ESE site. Potential alteration of splicing. | Benign/Little Clinical Significance | 1-Not pathogenic or of no clinical significance | 1-Neutral | Benign | Benign | 22 |
| 11 | c.3119G>A | p.Ser1040Asn | S1040N | 3238G>A | M | 17: 41244429 | rs4986852 | 0.00978 (T) | 0.00978 (T) | 0,00978 | − | 0,01109 | 0,01571 | 0.035304 | Tolerated (0.21) | Possibly Damaging (0.831) | Neutral | Class C0 | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Benign/Little Clinical Significance | 1-Not pathogenic or of no clinical significance | 1-Neutral | Benign | Benign | 5 |
| 11 | c.3305A>G | p.Asn1102Ser | N1102S | − | M | 17: 41244243 | rs80356900 | − | 0,00002 | − | 0,00008 | 0,00001 | 0,00001 | − | Tolerated (0.17) | Benign (0.156) | Deleterious | Class C0 | Creation of an exonic ESS site. Potential alteration of splicing. | Not Yet Reviewed | 2-Likely not pathogenic or of little clinical significance | 3-UV | Uncertain significance? | Uncertain Significance | 1 |
| 11 | c.3548A>G | p.Lys1183Arg | K1183R | 3667A>G | M | 17: 41244000 | rs16942 | 0.35264 (C) | 0,34901 | 0,35264 | 0,29525 | 0,31548 | 0,30133 | 0.299672 | Tolerated (1) | Benign (0) | Neutral | Class C0 | Creation of an exonic ESS site. Alteration of an exonic ESE site. Potential alteration of splicing. | Benign/Little Clinical Significance | 1-Not pathogenic or of no clinical significance | 1-Neutral | Benign | Benign | 22 |
| 11 | c.3752G>A | p.Cys1251Tyr | C1251Y | − | M | 17: 41243796 | rs879254079 | − | − | − | − | − | − | − | Tolerated (1) | Benign (0.001) | Neutral | Class C0 | Alteration of an exonic ESE site. Potential alteration of splicing. | Not Yet Reviewed | ND | ND | Uncertain significance? | Uncertain Significance | 1 |
| 11 | c.4039A>G | p.Arg1347Gly | R1347G | 4158A>G | M | 17: 41243509 | rs28897689 | 0.00060 (C) | 0.00398 | 0,0006 | 0,00484 | 0,00423 | 0,00481 | 0.005747 | Tolerated (0.09) | Benign (0.071) | Neutral | Class C0 | Creation of an exonic ESS site. Alteration of an exonic ESE site. Potential alteration of splicing. | Benign/Little Clinical Significance | 1-Not pathogenic or of no clinical significance | 1-Neutral | Benign | Benign | 2 |
| 12 | c.4113G>A | p.Gly1371= | G1371= | − | Syn | 17: 41243033 | rs147448807 | 0.00160 (T) | 0.00050 | 0,0016 | 0,00123 | 0,00156 | − | − | − | − | Neutral | − | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Likely benign | ND | 2-Likely Neutral | Likely Benign | Likely Benign | 1 |
| 13 | c.4308T>C | p.Ser1436= | S1436= | 4427T>C | Syn | 17: 41234470 | rs1060915 | 0.33626 (G) | 0,3431 | 0,33626 | 0,27956 | 0,30142 | 0,28489 | 0.283251 | − | − | Neutral | − | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 22 |
| 15 | c.4485-63C>G | − | − | IVS 14-63C>G | IVS | 17: 41226601 | rs273900734 | 0.35344 (C) | − | 0,35344 | − | 0,31645 | 0,30211 | 0.300493 | − | − | − | − | − | Benign/Little Clinical Significance | ND | ND | Benign | Benign | 2 |
| 16 | c.4837A>G | p.Ser1613Gly | S1613G | 4956A>G | M | 17: 41223094 | rs1799966 | 0.35583 (C) | − | 0,35583 | 0,29817 | − | 0,30333 | 0.300987 | Tolerated (0.11) | Benign (0.038) | Neutral | Class C0 | Alteration of an exonic ESE site. Potential alteration of splicing. | Benign/Little Clinical Significance | 1-Not pathogenic or of no clinical significance | 1-Neutral | Benign | Benign | 22 |
| 16 | c.4987-92A>G | − | − | IVS16-92A>G | IVS | 17: 41219804 | rs8176233 | 0.35463 (C) | − | 0,35463 | − | 0,31276 | 0,30294 | 0.300493 | − | − | − | − | Alteration of an exonic ESE site. Potential alteration of splicing.- | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 3 |
| 16 | c.4987-68A>G | − | − | VS16-68A>G | IVS | 17: 41219780 | rs8176234 | 0.35463 (C) | − | 0,35463 | − | 0,31483 | 0,30295 | 0.301314 | − | − | − | − | Alteration of an exonic ESE site. Potential alteration of splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 3 |
| 17 | c.5074+2T>C | − | − | IVS17+2T>C | SS | 17: 41219623 | rs80358089 | − | − | − | − | − | − | − | − | − | − | − | Alteration of the WT donor site, most probably affecting splicing | Pathogenic | 5-Definitely pathogenic | ND | Pathogenic? | Pathogenic | 1 |
| 17 | c.5075-53C>T | − | − | IVS17-53C>T | IVS | 17: 41216021 | rs8176258 | 0.01098 (A) | − | 0,01098 | 0,01708 | 0,01825 | 0,01721 | 0.018062 | − | − | − | − | Alteration of an intronic ESS site. Probably no impact on splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 2 |
| 18 | c.5123C>A | p.Ala1708Glu | A1708E | 5242C>A | M | 17: 41215920 | rs28897696 | − | 0,02487 | − | 0.00023 (T) | − | − | − | Deleterious (0) | Possibly Damaging (0.633) | Neutral | Class C65 | Activation of an exonic cryptic acceptor site, with presence of one or more cryptic branch point(s). Creation of an exonic ESS site. Alteration of an exonic ESE site. Potential alteration of splicing. | Pathogenic | 5-Definitely pathogenic | 5-Causal | Pathogenic? | Pathogenic? | 1 |
| 18 | c.5152+66G>A | − | − | IVS18+66G>A | IVS | 17: 41215825 | rs3092994 | 0.34245 (T) | − | 0,34245 | − | 0,31394 | 0,29599 | 0.291461 | − | − | − | − | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 3 |
| 21 | c.5304C>T | p.Cys1768= | C1769C | − | M | 17: 41203108 | rs138493864 | 0.00060 (A) | 0.00002 | 0,0006 | 0,00015 | 0,00013 | 0,00009 | − | − | − | Neutral | − | Creation of an exonic ESS site. Potential alteration of splicing. | Likely benign | ND | ND | Likely Benign | Likely Benign | 1 |
HGVS: Human Genome Variation Society; MAF: Minor allele frequency; ESP: NHLBI Exome Sequencing Project Exome Variant Server; gnomAD: The Genome Aggregation Database; TOPMed: Trans-Omics for Precision Medicine; ABraOM: Brazilian genomic variants; SIFT: Sorting intolerant from tolerant; PolyPhen: Polymorphism Phenotyping; Provean: Protein Variation Effect Analyzer; Align-GVGD: Class C0 (less probable to interfere with protein function), C15, C25, C35, C45, C55, C65 (more probable to interfere with protein function); Syn: Synonymous; IVS: Intervening sequence; M: Missense; SS: splice site; ND, Not determined; n: Number of patients harboring the variant
BRCA2 variants.
| Exon | HGVS Nucleotide | HGVS Protein | Protein Abbrev | Other names | Type | Localization (GRCh37) | NCBI 1000 Genomes Browser | Global MAF dbSNP | Allele Frequency ExAC | Global MAF 1000 genomes | ESP | gnomAD | TOPMed | ABraOM | SIFT | PolyPhen | Provean | Align-GVGD (Pufferfish) | Human Splicing Finder | BRCA Exchange | BRCA Mutation Database | BRCA Share™ | ClinVar | Interpretation | n |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | c.-26G>A | − | − | 203G>A | 5'UTR | 13: 32890572 | rs1799943 | 0.20927 (A) | 0,24652 | 0,20927 | 0,20883 | 0,22032 | 0,21567 | 0.217570 | − | − | − | − | ND | Benign/Little Clinical Significance | ND | ND | Benign | Benign | 21 |
| 2 | c.-15A>C | − | − | 214A>C | 5'UTR | 13: 32890583 | rs138705202 | 0.00080 (C) | 0.00022 | 0,0008 | 0,00038 | 0,00064 | 0,00076 | 0.002463 | − | − | − | − | ND | Not Yet Reviewed | ND | ND | Benign/Likely Benign | Likely benign | 1 |
| 2 | c.-11C>T | − | − | 218C>T | 5'UTR | 13: 32890587 | rs76874770 | 0.00439 (T) | 0,00163 | 0,00439 | 0,00584 | 0,0051 | 0,00546 | 0.007389 | − | − | − | − | ND | Benign/Little Clinical Significance | ND | ND | Benign | Benign | 2 |
| 2 | c.2T>G | p.Met1Arg | M1R | − | M | 13: 32890599 | rs80358547 | − | 0,00001 | − | − | 0,00001 | − | − | Damaging (0.00) | Probably Damaging (0.998) | Deleterious | Class C65 | ND | Not Yet Reviewed | 5-Definitely pathogenic | 5-Causal | Pathogenic? | Pathogenic | 1 |
| 3 | c.125A>G | p.Tyr42Cys | Y42C | 353A>G | M | 13: 32893271 | rs4987046 | 0.00080 (G) | 0,0017 | 0,0008 | 0,00246 | 0,00162 | 0,00158 | 0.001642 | Tolerate (0.12) | Benign (0.090) | Neutral | Class C0 | Activation of an exonic cryptic donor site. Potential alteration of splicing. | Benign/Little Clinical Significance | 1-Not pathogenic or of no clinical significance | 1-Neutral | Benign | Benign | 1 |
| 4 | c.425+33A>G | − | − | IVS4+33A>C | IVS | 13: 32899354 | rs200065709 | 0.00060 (G) | 0,00052 | 0,0006 | 0,00031 | 0.00010 | 0,00029 | 0.000821 | − | − | − | − | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Not Yet Reviewed | ND | 2-Likely Neutral | Benign/Likely Benign | Likely benign | 1 |
| 4 | c.425+67A>C | − | − | IVS4+67A>C | IVS | 13: 32899388 | rs11571610 | 0.07428 (C) | − | 0,07428 | − | 0,03064 | 0,03973 | 0.045156 | − | − | − | − | Alteration of an intronic ESS site. Probably no impact on splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 6 |
| 6 | c.517-19C>T | − | − | IVS6-19C>T | IVS | 13: 32900617 | rs11571623 | 0.00819 (T) | 0,00219 | 0,00819 | 0,00738 | 0,00586 | 0,007 | 0.003284 | − | − | − | − | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 2 |
| 8 | c.681+56C>T | − | − | IVS8+56C>T | IVS | 13: 32903685 | rs2126042 | 0.18590 (T) | − | 0,1859 | − | 0,21627 | 0,20076 | 0.184729 | − | − | − | − | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Not Yet Reviewed | ND | 1-Neutral | Benign | Benign | 17 |
| 10 | c.865A>C | p.Asn289His | N289H | 1093A>C | M | 13: 32906480 | rs766173 | 0.07368 (C) | − | 0,07368 | 0,03055 | 0,03055 | 0,03968 | 0.045156 | Damaging (0.003) | Benign (0.278) | Neutral | Class C0 | Alteration of an exonic ESE site. Potential alteration of splicing | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 7 |
| 10 | c.1114A>C | p.His372Asn | H372N | 1342 A>C | M | 13: 32906729 | rs144848 | 0.24940 (C) | 0,27793 | 0,2494 | − | 0,22303 | 0,23657 | 0.259442 | Tolerated (0.35) | Benign (0.00) | Neutral | Class C0 | Alteration of an exonic ESE site. | Benign/Little Clinical Significance | 1-Not pathogenic or of no clinical significance | 1-Neutral | Benign | Benign | 19 |
| 10 | c.1365A>G | p.Ser455= | S455= | 1593A>G | Syn | 13: 32906980 | rs1801439 | 0.07368 (G) | 0,05178 | 7368 | 0,03101 | 0,03048 | 0,03968 | 0.045156 | − | − | Neutral | − | Alteration of an exonic ESE site. Potential alteration of splicing | ND | ND | 1-Neutral | Benign | Benign | 7 |
| 10 | c.1514T>C | p.Ile505Thr | I505T | M | 13: 32907129 | rs28897708 | 0.00040 (C) | 0,00072 | 0,0004 | 0,00077 | 0,00083 | 0,00065 | 0.000821 | Tolerated (0.1) | Possibly Damaging (0.651) | Neutral | Class C0 | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Benign/Little Clinical Significance | 1-Not pathogenic or of no clinical significance | 1-Neutral | Benign | Benign | 1 | |
| 10 | c.1909+92_1909+96del | − | − | IVS10+92del5 | IVS | 13: 32907615-32907620 | rs144549870 | 0.01577 (TAT) | − | − | − | − | − | 0.006568 | − | − | − | − | − | ND | ND | ND | Benign | Benign | 2 |
| 10 | c.1910-74T>C | − | − | IVS10-74T>C | IVS | 13: 32910328 | rs2320236 | 0.17452 (C) | − | 0,17452 | − | 0,20561 | 0,20561 | rs2320236 | − | − | − | − | Creation of an intronic ESE site. Probably no impact on splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 14 |
| 10 | c.1910-51G>T | − | − | IVS10-51G>T | IVS | 13: 32910351 | rs11571651 | 0.07348 (T) | 0,04934 | 0,07348 | 0,03056 | 0,03041 | 0,03968 | 0.045977 | − | − | − | − | Alteration of an intronic ESS site. Probably no impact on splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 6 |
| 11 | c.2229T>C | p.His743= | H743= | 2457T>C | Syn | 13: 32910721 | rs1801499 | 0.07348 (C) | 0,05158 | 0.07348 | 0,03129 | 0,03065 | 0,03972 | 0.045156 | − | − | Neutral | − | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 7 |
| 11 | c.2350A>G | p.Met784Val | M784V | 2578A>G | M | 13: 32910842 | rs11571653 | 0.00359 (G) | 0,00031 | 0,00359 | − | 0,00023 | 0,00022 | 0.002463 | Tolerated (1.00) | Benign (0.00) | Neutral | Class C0 | Creation of an exonic ESS site. Potential alteration of splicing. | Benign/Little Clinical Significance | 3-Uncertain | 3-UV | Benign | Benign | 1 |
| 11 | c.2971A>G | p.Asn991Asp | N991D | 3199A>G | M | 13: 32911463 | rs1799944 | 0.08007 (G) | 0,05341 | 0,08007 | 0,03725 | 0,03723 | 0,0461 | 0.046798 | Tolerated (1.00) | Benign (0.00) | Neutral | Class C0 | Alteration of an exonic ESE site. Potential alteration of splicing | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 7 |
| 11 | c.3264T>C | p.Pro1088= | P1088= | 3492T>C | Syn | 13: 32911756 | rs36060526 | 0.00679 (C) | 0.00238 | 0,00679 | 0,00756 | 0,00762 | 0,00756 | 0.006568 | − | − | Neutral | − | ND | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 1 |
| 11 | c.3371A>G | p.Gln1124Arg | Q1124R | M | 13: 32911863 | rs1555283204 | − | − | − | − | − | − | − | Damaging (0.01) | Probably Damaging (1.00) | Deleterious | Class C35 | Activation of an exonic cryptic donor site. Potential alteration of splicing. | Not Yet Reviewed | ND | ND | Uncertain significance | Uncertain significance | 1 | |
| 11 | c.3396A>G | p.Lys1132= | L1132= | 3624A>G | Syn | 13: 32911888 | rs1801406 | 0.26677 (G) | 0,29449 | 0,26677 | 0,27984 | 0,29762 | 0,28221 | 0.283251 | − | − | Neutral | − | Alteration of an exonic ESE site. Potential alteration of splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 23 |
| 11 | c.3807T>C | p.Val1269= | V1269= | 4035T>C | Syn | 13: 32912299 | rs543304 | 0.16813 (C) | 0,18985 | 0,16813 | 0,19111 | 0,18144 | 0,18622 | 0.187192 | − | − | Neutral | − | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 23 |
| 11 | c.4068G>A | p.Leu1356= | L1356= | 4296G>A | Syn | 13: 32912560 | rs28897724 | 0.00040 (A) | 0,00305 | 0,0004 | 0,00315 | 0.00245 | 0,00312 | 0.002463 | − | − | Neutral | − | ND | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 1 |
| 11 | c.4090A>C | p.Ile1364Leu | I1364L | 4318A>C | M | 13: 32912582 | rs56248502 | 0.00439 (C) | 0,00172 | 0,00439 | 0,00631 | 0,00577 | 0.006568 | Tolerated (0.76) | Benign (0.001) | Neutral | Class C0 | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 2 | |
| 11 | c.4258G>T | p.Asp1420Tyr | D1420Y | 4486G>T | M | 13: 32912750 | rs28897727 | 0.00399 (T) | 0,0068 | 0,00399 | 0,00396 | 0,00794 | 0,00425 | 0.001642 | Damaging (0.01) | Benign (0.030) | Deleterious | Class C15 | ND | Benign/Little Clinical Significance | 1-Not pathogenic or of no clinical significance | 1-Neutral | Benign | Benign | 1 |
| 11 | c.5418A>G | p.Glu1806= | E1806= | 5646A>G | Syn | 13: 32913910 | rs34351119 | 0.00679 (G) | 0,00233 | 0,00679 | 0,0083 | 0,00764 | 0,00785 | 0.006568 | − | − | Neutral | − | ND | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 1 |
| 11 | c.5640T>G | p.Asn1880Lys | N1880K | 5868T>G | M | 13: 32914132 | rs11571657 | 0.00220 (G) | 0,00076 | 0,0022 | 0,00315 | 0,00264 | 0,00294 | 0.000821 | Damaging (0.05) | Benign (0.167) | Neutral | Class C0 | Creation of an exonic ESS site. Potential alteration of splicing. | Benign/Little Clinical Significance | 2-Likely not pathogenic or of little clinical significance | 2-Likely Neutral | Benign/Likely Benign | Likely benign | 1 |
| 11 | c.5645C>A | p.Ser1882Ter | S1882X | 5873C>A | N | 13: 32914137 | rs80358785 | − | 0,00002 | − | − | 0,00002 | 0,00002 | − | − | − | − | − | Alteration of an exonic ESE site. Potential alteration of splicing. | Pathogenic | 5-Definitely pathogenic | 5-Causal | Pathogenic | Pathogenic | 1 |
| 11 | c.5744C>T | p.Thr1915Met | T1915M | 5972C>T | M | 13: 32914236 | rs4987117 | 0.00859 (T) | 0,02114 | 0,02114 | 0,02114 | 0.00859 (T) | 0,01744 | 0.017241 | Tolerated (0.13) | Benign (0.000) | Neutral | Class C0 | Creation of an exonic ESS site. Potential alteration of splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 2 |
| 11 | c.5768A>C | p.Asp1923Ala | D1923A | 5996A>C | M | 13: 32914260 | rs45491005 | 0.00020 (C) | − | 0,0002 | 0,0002 | 0.00054 | 0.00105 | − | Tolerated (0.29) | Benign (0.144) | Deleterious | Class C0 | Alteration of an exonic ESE site. Potential alteration of splicing. | Benign/Little Clinical Significance | 2-Likely not pathogenic or of little clinical significance | 2-Likely Neutral | Benign | Likely benign | 1 |
| 11 | c.6841+53delTATTCAGTAG | − | − | − | IVS | 13: 32915384-32915394 | − | − | − | − | − | − | − | − | − | − | − | − | Alteration of an intronic ESS site. Probably no impact on splicing. | ND | ND | ND | ND | Uncertain Significance | 1 |
| 11 | c.6841+80delTTAA | − | − | IVS11+80delTTAA | IVS | 13: 32915411-32915414 | rs11571661 | 0.26578 (AA) | − | − | − | − | − | 0.279605 | − | − | − | − | Creation of an intronic ESE site. Probably no impact on splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 7 |
| 14 | c.7017G>C | p.Lys2339Asn | K2339N | 7245 G>C | M | 13: 32929007 | rs45574331 | 0.00679 (C) | 0,00228 | 0,00679 | 0,00808 | 0,00764 | 0,00786 | 0.006568 | Damaging (0.01) | Benign (0.105) | Neutral | Class C0 | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Benign/Little Clinical Significance | ND | 2-Likely Neutral | Benign | Benign | 1 |
| 14 | c.7242A>G | p.Ser2414= | S2114= | 7470A>G | Syn | 13: 32929232 | rs1799955 | 0.23263 (G) | − | 0,23263 | 0,21136 | − | 0,22464 | 0.238095 | − | − | Neutral | − | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 19 |
| 14 | c.7319A>G | p.His2440Arg | H2440R | 7547A>G | M | 13: 32929309 | rs4986860 | 0.01038 (G) | 0,00304 | 0,01038 | 0,01054 | 0,00946 | 0,00967 | 0.007389 | Tolerated (0.55) | Benign (0.002) | Neutral | Class C0 | ND | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 1 |
| 14 | c.7397T>C | p.Ala2466Val | A2466V | − | M | 13: 32929387 | rs169547 | 0.02416 (T) | 0,99372 | 0.97584 | 0,9777 | 0,97881 | 0,98191 | 0.983580 | Tolerated (0.98) | Possibly Damaging (0.793) | Neutral | Class C0 | ND | ND | ND | 1-Neutral | Benign | Benign | 50 |
| 14 | c.7435+53C>T | − | − | IVS14+53C>T | IVS | 13: 32929478 | rs11147489 | 0.07248 (T) | − | 0,07248 | − | 0,0301 | 0,03924 | − | − | − | − | − | Creation of an intronic ESE site. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 1 |
| 15 | c.7469T>C | p.Ile2490Thr | I2490T | 7697T>C | M | 13: 32930598 | rs11571707 | 0.01597 (C) | 0,01436 | 0.01597 | 0,00161 | 0,0035 | 0,00913 | 0.021346 | Tolerated (1.00) | Benign (0.010) | Neutral | Class C45 | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 1 |
| 17 | c.7806-14T>C | − | − | IVS16-14T>C | IVS | 13: 32936646 | rs9534262 | 0.46845 (T) | 0,52083 | 0,53155 | 0,52015 | 0,54679 | 0,53151 | 0.523810 | − | − | − | − | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Benign/Little Clinical Significance | ND | 3-UV | Benign | Likely benign | 36 |
| 19 | c.8460A>C | p.Val2820= | V2820= | 8688A>C | Syn | 13: 32944667 | rs9590940 | 0.01438 (C) | 0,00368 | 0,01438 | 0,01299 | 0,01105 | 0,01219 | 0.006568 | − | − | Neutral | − | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 2 |
| 19 | c.8487+47C>T | − | − | IVS19+47C>T | IVS | 13: 32944741 | rs11571744 | 0.01617 (T) | − | 0,01617 | 0,01523 | − | 0,01481 | 0.006568 | − | − | − | − | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Benign/Little Clinical Significance | ND | 3-UV | Benign | Benign | 3 |
| 20 | c.8632+132dup | − | − | c.IVS20+132insC | IVS | 13: 32945368-32945369 | rs201392123 | 0.00899 (CC) | − | 0.00899 | − | 0,00619 | 0,00754 | 0.002627 | − | − | − | − | ND | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 2 |
| 21 | c.8755-66T>C | − | − | IVS21-66T>C | IVS | 13: 32953388 | rs4942486 | 0.48842 (T) | − | 0,51158 | − | 0,52569 | 0,51037 | 0.508210 | − | − | − | − | Alteration of an intronic ESS site. Probably no impact on splicing. Creation of an intronic ESE site. Probably no impact on splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 38 |
| 22 | c.8851G>A | p.Ala2951Thr | A2951T | 9079G>A | M | 13: 32953550 | rs11571769 | 0.00998 (A) | 0,00785 | 0.00998 | 0,00438 | 0,00363 | 0,00721 | 0.013136 | Damaging (0.00) | Probably Damaging (1.00) | Neutral | Class C55 | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 1 |
| 22 | c.8942A>G | p.Glu2981Gly | E2981G | 9170A>G | M | 13: 32953641 | rs398122716 | − | 0,00001 | − | − | 0,00002 | 0,00001 | − | Tolerated (0.16) | Benign (0.030) | Neutral | Class C65 | ND | ND | ND | 3-UV | Conflicting interpretations of pathogenicity? Likely benign(1);Uncertain significance(3) | Uncertain significance | 1 |
| 23 | c.9038C>T | p.Thr3013Ile | T3013I | − | M | 13: 32953971 | rs28897755 | − | 0,00023 | − | 0,00046 | 0,00019 | 0,0002 | − | Tolerated (0.24) | Probably Damaging (0.875) | Neutral | Class C0 | ND | Benign/Little Clinical Significance | 1-Not pathogenic or of no clinical significance | 1-Neutral | Benign | Benign | 1 |
| 24 | c.9257-83G>A | − | − | IVS24-83G>A | IVS | 13: 32968743 | rs9595456 | 0.05052 (A) | − | 0,05052 | − | 0,04116 | 0.04575 | 0.022989 | − | − | − | − | Creation of an intronic ESE site. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 4 |
| 24 | c.9257-16T>C | − | − | IVS24-16T>C | IVS | 13: 32968810 | rs11571818 | 0.00439 (C) | 0,00439 | 0,00765 | 0,00592 | 0,00548 | 0,00548 | 0.004926 | − | − | − | − | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | ND | ND | 3-UV | Benign | Likely benign | 1 |
| 27 | c.9730G>A | p.Val3244Ile | V3244I | 9958 G>A | M | 13: 32972380 | rs11571831 | 0.00679 (A) | − | − | 0,0083 | 0,00767 | 0,00787 | − | Tolerated (0.49) | Benign (0.000) | Neutral | Class C0 | ND | Benign/Little Clinical Significance | ND | 2-Likely Neutral | Benign | Benign | 1 |
| 27 | c.9976A>T | p.Lys3326Ter | K3326X | 10204A>T | N | 13: 32972626 | rs11571833 | 0.00439 (T) | 0,00702 | 0,00439 | 0.00646 | 0,00544 | 0,00547 | 0.004926 | − | − | − | − | Creation of an exonic ESS site. Potential alteration of splicing. Alteration of an exonic ESE site. Potential alteration of splicing. | Benign/Little Clinical Significance | 2-Likely not pathogenic or of little clinical significance | 1-Neutral | Benign | Benign | 1 |
| 27 | c.10110G>A | p.Arg3370= | R3370= | − | Syn | 13: 32972760 | rs28897762 | 0.00080 (A) | 0,00147 | 0,0008 | 0,00215 | 0,0014 | 0,00131 | 0.000821 | − | − | Neutral | − | Alteration of an exonic ESE site. Potential alteration of splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 1 |
| 27 | c.10234A>G | p.Ile3412Val | I3412V | 10462 A>G | M | 13: 32972884 | rs1801426 | 0.04493 (G) | 0,02266 | 0,04493 | 0,03729 | 0,0369 | 0,04054 | 0.021346 | Tolerrated (0.34) | Benign (0.002) | Neutral | Class C0 | Alteration of an exonic ESE site. Potential alteration of splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 4 |
HGVS: Human Genome Variation Society; MAF: Minor allele frequency; EXAC: Exome Aggregation Consortium; ESP: NHLBI Exome Sequencing Project Exome Variant Server; gnomAD: The Genome Aggregation Database; TOPMed: Trans-Omics for Precision Medicine; ABraOM: Brazilian genomic variants; SIFT: Sorting Intolerant From Tolerant; PolyPhen: Polymorphism Phenotyping; Provean: Protein Variation Effect Analyzer; Align-GVGD: Class C0 (less probable to interfere with protein function), C15, C25, C35, C45, C55, C65 (more probable to interfere with protein function); Syn: Synonymous; IVS: Intervening sequence; M: Missense; SS: Splice site; ND: Not determined; n: Number of patients bearing the variant
Mutations in BRCA1 comprised one splice-site variant (c.5074+2T>C, in exon 17), one missense mutation (c.5123C>A), and one BRCA1 rearrangement generating a large deletion encompassing exons 1-19. The two pathogenic mutations in BRCA2 included one missense variant (c.2T>G) and one nonsense variant (c.5645C>A) (Table 2). The presence of CHEK2 c.1100delC mutation was investigated in 47 out of the 49 patients; however, no mutations were detected.
Eight VUS were detected, five in BRCA1 and three in BRCA2. Among the VUS, four distinct missense variants were identified, two in each gene (BRCA1: c.3305A>G and c.3752G>A; BRCA2: c.3371A>G and c.8942A>G), among which BRCA2 c.3371A>G was predicted to be deleterious by at least three out of four mutation function prediction models (SIFT, Polyphen-2, Align-GVGD, or Provean) (Table 3). The remaining VUS were located in the intronic regions, at least 36 nucleotides away from the intron-exon boundary.
In silico analysis of VUS identified in BRCA1 and BRCA2 using mutation function prediction models.
| Gene | HDVS cDNA | HGVS protein | SIFT | PolyPhen | Align-GVGD | Provean | Human Splicing Finder | ID |
|---|---|---|---|---|---|---|---|---|
| BRCA1 | c.3305A>G | p.Asn1102Ser | Tolerated | Benign | Class C0 | Deleterious | Creation of an exonic ESS site. Potential alteration of splicing. | 49 |
| c.3752G>A | p.Cys1251Tyr | Tolerated | Benign | Class C0 | Neutral | Alteration of an exonic ESE site. Potential alteration of splicing. | 48 | |
| BRCA2 | c.3371A>G | p.Gln1124Arg | Damaging | Probably Damaging | Class C35 | Deleterious | Activation of an exonic cryptic donor site. Potential alteration of splicing. | 24 |
| c.8942A>G | p.Glu2981Gly | Tolerated | Benign | Class C65 | Neutral | ND | 12 |
Tumor sequencing was performed on samples from 27 elderly patients to identify PIK3CA mutations. Fourteen tumors (51.8%) were found to harbor mutations in exons 9 or 20; however, only ten (37%) harbored meaningful deleterious or possibly deleterious variants (pathogenic in at least one out of four function prediction tests). Recurrent mutations were E545A (observed in four samples) and H1047L (in the other two samples). Among these 27 elderly patients, two were BRCA1 mutation carriers, both of whom harbored somatic pathogenic (E545A) or possibly pathogenic PIK3CA mutations (Additional Table 4).
In silico analysis of the alterations in exons 9 and 20 of PIK3CA in postmenopausal patients with breast cancer.
| Sample ID | Age at diagnosis | Molecular Subtype | Exon | Cdna | Protein | Protein | Mutation Type | ID COSMIC | Polyphen | SIFT | Provean | Align-GVGD |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 71 | Luminal B | 9 | c.1634A>C | p.Glu545Ala | E545A | M | COSM12458 | Probably Damaging | Damaging | Deleterious | Class C65 |
| 3 | 66 | Luminal B | 9 | c.1639G>C | p.Glu547Gln | E547Q | M | − | Probably Damaging | Damaging | Neutral | Class C25 |
| 8 | 61 | Luminal B | 20 | c.3075C>T | p.Thr1025= | T1025T | Syn | COSM21451 | − | Tolerated | Neutral | − |
| c.3140A>T | p.His1047Leu | H1047L | M | COSM776 | Benign | Damaging | Neutral | Class C65 | ||||
| 9 | 73 | Luminal A | 9 | c.1629C>T | p.Ile543= | I543I | Syn | COSM5020257 | − | Tolerated | Neutral | − |
| 10 | 57 | Luminal A | 9 | c.1549C>T | p.Leu517= | L517L | Syn | − | − | Tolerated | Neutral | − |
| 17* | 56 | Luminal B | 9 | c.1634A>C | p.Glu545Ala | E545A | M | COSM12458 | Probably Damaging | Damaging | Deleterious | Class C65 |
| 21 | 67 | Luminal B | 9 | c.1550T>C | p.Leu517Pro | L517P | M | − | Benign | Damaging | Neutral | Class C65 |
| 23 | 62 | Luminal B | 20 | c.3140A>T | p.His1047Leu | H1047L | M | COSM776 | Benign | Damaging | Neutral | Class C65 |
| 26 | 60 | Luminal B | 9 | c.1547G>A | p.Arg516Lys | R516K | M | COSM3724545 | Benign | Tolerated | Neutral | Class C25 |
| 20 | c.3170G>A | p.Trp1057* | W1057X | N | COSM6475611 | − | − | − | − | |||
| 32 | 56 | Luminal A | 20 | c.3098A>G | Gln1033Arg | Q1033R | M | COSM303947 | Possible Damaging | Damaging | Neutral | Class C35 |
| 36 | 63 | Luminal A | 9 | c.1634A>C | p.Glu545Ala | E545A | M | COSM12458 | Probably Damaging | Damaging | Deleterious | Class C65 |
| c.1658_1659delGTinsC | p.Ser553Thrfs*7 | S553fs | F | − | − | − | − | − | ||||
| 39 | 63 | Luminal A | 9 | c.1638G>A | p.Gln546= | Q546Q | Syn | COSM5622324 | − | Toleratd | Neutral | − |
| c.1664+46G>A | − | − | IVS | − | − | − | − | − | ||||
| 20 | c.3102G>A | p.Glu1034= | E1034E | Syn | − | − | Tolerated | Neutral | − | |||
| 46 | 55 | HER2 | 9 | c.1634A>C | p.Glu545Ala | E545A | M | COSM12458 | Probably Damaging | Damaging | Deleterious | Class C65 |
| c.1651C>T | p.Leu551= | L551L | Syn | COSM308546 | − | Tolerated | Neutral | − | ||||
| c.1658_1659delGTinsC | p.Ser553Thrfs*7 | S553fs | F | − | − | − | − | − | ||||
| 47* | 58 | TN | 9 | c.1622C>T | p.Ser541Phe | S541F | M | COSM6438100 | Possible Damaging | Damaging | Deleterious | Class C65 |
| 20 | c.3110A>T | p.Glu1037Val | E1037V | M | − | Benign | Damaging | Deleterious | Class C65 |
HGVS: Human Genome Variation Society; SIFT: Sorting Intolerant From Tolerant; PolyPhen: Polymorphism Phenotyping; Provean: Protein Variation Effect Analyzer; Align-GVGD: Class C0 (less probable to interfere with protein function), C15, C25, C35, C45, C55, C65 (more probable to interfere with protein function); Syn: Synonymous; IVS: Intervening Sequence; M: Missense; N: Nonsense. *Patients also harboring pathogenic germline mutations in BRCA1.
Another three tumors (11.1%), all luminal A subtypes, harbored synonymous variants (in one case, associated with an intronic variant) (sample 39). In addition, tumors from another six patients harbored multiple PIK3CA variants; however, two tumors harbored (samples 26 and 39) a combination of non-pathogenic variants represented by missense non-pathogenic and nonsense variants (sample 26) or a combination of a deep intronic and two synonymous variants (sample 39). In the third tumor, PIK3CA double mutation (sample 47) (S541P and E1037V) was considered pathogenic in at least three function prediction tests, even though none of them were located in a hotspot. In the fourth and fifth tumors (samples 36 and 46), the contribution of the mutations were difficult to define because the PIK3CA pathogenic missense variant (E545A) was accompanied by a frameshift (FS) mutation (S553FS). If it occurs in cis, FS S553FS might counteract the oncogenic potential of E545A. The sixth tumor (sample 8) harbored a pathogenic hotspot (H1047L) and a synonymous variant (Additional Table 4).
In a cohort of young patients, PIK3CA variants were observed in 12 tumors, including synonymous variants—detected in two tumors (one luminal B, sample 484, and one HER2+ sample 503)—and missense non-pathogenic variants detected in another two samples (samples 455 and 478). In addition, a nonsense variant, W552* was detected in a luminal A tumor (sample 468). Hence, pathogenic or possibly pathogenic PIK3CA mutations were detected in seven out of forty-one young patients (17.1%) (Additional Table 5).
In silico analysis of the alterations in exons 9 and 20 of PIK3CAin young patients with breast cancer.
| Sample ID | Age at diagnosis | Molecular Subtype | Exon | cDNA | Protein | p.Asn1044Asp | Mutation Type | ID COSMIC | Polyphen | SIFT | Provean | Align-GVGD |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 452 | 34 | Luminal B | 20 | c.3130A>G | p.Asn1044Asp | N1044D | M | COSM27134 | Probably Damaging | Tolerated | Neutral | Class C15 |
| 454 | 34 | Luminal | 20 | c.3146G>A | p.Gly1049Asp | G1049D | M | COSM308548 | Probably Damaging | Tolerated | Neutral | Class C65 |
| 455 | 28 | Luminal B | 9 | c.1558G>A | p.Asp520Asn | D520N | M | COSM29096 | Benign | Tolerated | Neutral | Class C15 |
| 457 | 33 | Luminal B | 9 | c.1615C>T | p.Pro539Ser | P539S | M | COSM249880 | Probably Damaging | Tolerated | Deleterious | Class C65 |
| c.1664G>A | p.Arg555Lys | R555K | M | COSM1716158 | Probably Damaging | Damaging | Deleterious | Class C25 | ||||
| 468 | 33 | Luminal A | 9 | c.1656G>A | p.Trp552* | W552X | N | COSM37025 | − | − | − | − |
| 477 | 27 | TN | 9 | c.1634A>C | p.Glu545Ala | E545A | M | COSM12458 | Probably Damaging | Damaging | Deleterious | Class C65 |
| 478 | 29 | HER2 | 20 | c.3165G>A | p.Met1055Ile | M1055I | M | COSM9146166 | Benign | Tolerated | Neutral | Class C0 |
| c.3201G>A | p. Leu1067= | L1067L | Syn | − | − | Tolerated | Neutral | − | ||||
| 480 | 31 | TN | 9 | c.1634A>C | p.Glu545Ala | E545A | M | COSM12458 | Probably Damaging | Damaging | Deleterious | Class C65 |
| 20 | c.3203A>C | p.Asn1068Thr | N1068T | M | − | Probably Damaging | Damaging | Neutral | Class C55 | |||
| 483 | 35 | Luminal B | 9 | c.1634A>C | p.Glu545Ala | E545A | M | COSM12458 | Probably Damaging | Damaging | Deleterious | Class C65 |
| 484 | 35 | Luminal B | 9 | c.1593C>A | p.Leu531= | L531L | Syn | − | − | Tolerated | Neutral | − |
| 503 | 35 | HER2 | 20 | c.3150C>T | p.Gly1050= | G1050G | Syn | − | − | Tolerated | Neutral | − |
| 518 | 31 | Luminal B | 9 | c.1615C>T | p.Pro539Ser | P539S | M | COSM249880 | Probably Damaging | Tolerated | Deleterious | Class C65 |
HGVS: Human Genome Variation Society; SIFT: Sorting Intolerant From Tolerant; PolyPhen: Polymorphism Phenotyping; Provean: Protein Variation Effect Analyzer; Align-GVGD: Class C0 (less probable to interfere with protein function), C15, C25, C35, C45, C55, C65 (more probable to interfere with protein function); Syn: Synonymous; M: Missense; N: Nonsense.
Among the young patients, E545A was the most frequent mutation (detected in three different samples, one luminal B and two triple-negative tumors). In one of these triple-negative tumors, E545A occurred concomitantly with N1068T, another pathogenic variant. The variant P539S, considered pathogenic in the prediction models, was detected in two luminal B samples, in one of these cases, in combination with R555K, which is also a pathogenic variant.
We then compared frequency of pathogenic PIK3CA mutation in tumors from postmenopausal and young patients (37% vs. 17%); however, we could not find a significant difference (Table 4). Using our data with a sample size of 27 postmenopausal and 41 young women and the reported frequency of PIK3CA mutation, the power to detect a difference with a one-sided significance level of 0.05% was 58.51%.
The frequency of PIK3CA is enriched in ER-positive tumors, and in a previous study we detected a trend toward a higher frequency of PIK3CA mutations in ER-positive tumor from elderly women compared to that observed in younger women (7). Upon considering the characteristics of the patients in the present series, we observed differences between the two groups, reflecting a higher proportion of luminal tumors in postmenopausal women. We then analyzed the frequency of PIK3CA mutations in luminal tumors and observed that eight out of the twenty-four samples (33.3%) from postmenopausal patients and five out of the twenty-five samples (20%) from young patients harbored pathogenic PIK3CA mutations (Table 4; p=0.291). A future meta-analysis including more recent data may help to clarify this aspect.
We next considered a total of 68 patients, postmenopausal as well as young, who were tested for the presence of germline mutations in BRCA1/BRCA2 and somatic mutations in PIK3CA. Upon simultaneously considering patients from both age groups, two out of eight germline BRCA1/BRCA2 mutant carriers (25%) were also found to harbor somatic mutations in PIK3CA. Among the 60 patients who were BRCA1 and BRCA2 wild type, 15 manifested tumors harboring PIK3CA mutations (25%).
DISCUSSIONIn this cohort of postmenopausal breast cancer patients, 10.2% harbored pathogenic germline BRCA1/BRCA2 variants; 11.7% of these patients had at least one family member who was affected with breast, ovarian, prostate, or pancreatic cancer.
Age at the onset of breast cancer and a family history of breast and ovarian cancer are important factors associated with the frequency of germline BRCA mutations (29). For elderly patients who were not selected for a family history of cancer, the frequency of BRCA mutations tended to be relatively low. Accordingly, a recent nested case-control study conducted in the USA revealed that only 1.18% of the unselected postmenopausal breast cancer patients were BRCA1/BRCA2 mutation carriers (27). In a large cohort comprising 1554 Brazilian breast cancer patients referred for genetic testing at a single clinical diagnostic laboratory in Brazil, 9.84% were found to be BRCA1 or BRCA2 mutation carriers independent of age (30). Higher BRCA mutation frequencies (varying from 15% to 22%) have been reported among young Brazilian breast cancer patients with ages up to 35 years (22,31,30). However, specifically for postmenopausal Brazilian patients with breast cancer, little data are available. Our study indicates that 10.6% of the breast cancer patients with at least one close relative affected by the disease (until third degree) harbor germline BRCA1/BRCA2 mutations. A previous study evaluated 39 breast cancer patients aged more than 50 years, among whom 17.9% were BRCA mutation carriers (32). These latter patients reported a strong family history based on the early age of cancer onset or multiple relatives with breast cancer and/or ovarian cancer at any age, which may explain the higher BRCA mutation frequency.
An important issue to take into consideration is the cost-effectiveness of the diagnostic program for germline mutations in BRCA1/BRCA2 genes and preventative strategies for relatives of patients diagnosed with the mutation. In the scenario of Brazilian ovarian cancer patients, for whom BRCA1/BRCA2 mutation frequency is 20%, performing genetic testing and adopting prophylactic measures for family members was considered a cost-effective measure (33). In a more inclusive model, BRCA testing may be offered to women of the general population to avoid missing mutation carriers, owing to test indications based on clinical criteria and family history. In this context, population-based BRCA testing was estimated to be cost-effective for the Brazilian population and to prevent a large number of breast and ovarian cancer cases (34). Although direct studies for postmenopausal Brazilian breast cancer patients are necessary, the previous two studies might suggest that genetic testing may be valuable for these women in the context of a positive family history.
The variants detected in the present study were not among the most frequent mutations in BRCA1 and BRCA2 in Brazilian patients with breast cancer. Variants BRCA1 c.5074+2T>C, BRCA1 c.5123C>A, and BRCA2 c.2T>G respectively represent 2.2%, 0.5%, and 1.2% of the BRCA1/BRCA2 mutations previously reported (28).
The other two BRCA mutations, BRCA1 large rearrangement (del exons 1-19) and BRCA2 c.5645C>A, have not been previously reported in the Brazilian population. The variant, BRCA2 c.5645C>A has been reported in breast cancer patients from Japan, China, and the Czech Republic (35,36,37), and in prostate cancer patients (38). Interestingly, our patient who harbored this variant was also born in Japan.
Somatic mutations in PIK3CA gene are the second most common mutations in breast cancer, just after TP53 (7). The PIK3CA mutation hotspots were clustered in exon 9 in nucleotides corresponding to codons E542K and E545K (helical domain) and in exon 20 in nucleotides corresponding to codon H1047R (kinase domain) (39,40).
In the present series, the most frequent mutation in PIK3CA in tumors from both postmenopausal and young patients was E545A, a variant with intermediate oncogenic potency, located in the helical domain (39). In agreement with our data, studies on breast cancer patients from Singapore and Peru have also found E545A to be the most frequent PIK3CA variant in tumor samples (41,42). Nevertheless, a method was developed to specifically enhance the detection of E454A (43). In contrast, data from another cohort of Brazilian patients with sporadic breast cancer have reported that the most frequent PIK3CA hotspot mutations were E542K, E545K, and H1047R (13).
The second most commonly found mutations in elderly patients were H1047L and S553FS. H1047L is located in the kinase domain and is associated with high oncogenic potential (39). Further, the frameshift mutation S553FS might counteract the proto-oncogene potential of PIK3CA. In addition, nonsense mutations were detected in tumors from both elderly and young patients, which might also neutralize the proto-oncogenic activity of PIK3CA. However, another study has specified that nonsense mutations in PIK3CA are not frequently encountered (44).
Six tumors were found to harbor double or triple PIK3CA variants (four from elderly patients and two from young patients). It has been previously shown that approximately 13% of all the PIK3CA mutations correspond to multiple variants occurring in the same tumor. It has also been reported that most double mutations occur in cis and induce the activation of the downstream PI3K pathway (compared to single-hotspot mutants) (40). However, in the present study, among the four tumors in elderly patients harboring double or triple variants, only one might be deleterious, involving a combination of S541P and E1037V. In the other three tumors, concomitant variants included nonsense, frameshift, synonymous, and intronic variants, in addition to missense variants with pathogenic or non-pathogenic potential. The determination of whether these variants were in cis might have helped to determine the oncogenic potential of the combinations because if a driver mutation occurred in trans, the effect of the driver mutation might have prevailed.
In the present cohort of patients, somatic mutations in PIK3CA were detected in 25% of the patients harboring germline BRCA1/BRCA2 mutations (two of the eight postmenopausal patients were analyzed for the presence of both gene mutations). This finding may be attributed to the small sample size. In other studies, the frequency of the combination of both mutations appeared to be less than that of individual mutations. In Chinese breast cancer patients, PIK3CA somatic mutations were detected in 14% and 43% of the patients harboring germline BRCA1/BRCA2 mutations (vs. wild type carriers), respectively (18). PIK3CA somatic mutations were not detected in male patients with breast cancer who harbored BRCA2 mutations (19).
Although we were not able to identify any associations between the germline BRCA and somatic PIK3CA mutations because of the small number of patients involved in this study, this is an intriguing situation involving two genes that are treatment targets; therefore, this information may be aggregated in future studies.
The limitations of our study are the small sample size and the sequencing of hotspots (but not all exons of PIK3CA), which may have resulted in the underestimation of the mutation frequency. The strengths of this study are the combined analysis of germline BRCA1/BRCA2 and somatic PIK3CA mutations in a group of postmenopausal and young patients with breast cancer.
In conclusion, the present data indicate that BRCA1/BRCA2 sequencing may be considered for postmenopausal breast cancer patients having a family history of cancer. In addition, although the frequency of PIK3CA variants in exons 9 and 20 is high in both elderly and young patients, some of these variants may not be pathogenic in the context of breast cancer.
AUTHOR CONTRIBUTIONSNagy TR conceived the study, enrolled patients, collected clinical data, performed the experiments, analyzed the data, analyzed and interpreted the mutational data, drafted the manuscript, and revised and approved the final version of the manuscript. Maistro S conceived the study, performed the experiments, analyzed the data, analyzed the mutational data, interpreted the data, drafted the manuscript, and revised and approved the final version of the manuscript. Encinas G conceived the study, performed the experiments, analyzed the mutational data, and revised and approved the final version of the manuscript. Katayama MLH performed the experiments, analyzed the data, analyzed the mutational data, interpreted the data, drafted the manuscript, and revised and approved the final version of the manuscript. Pereira GFL analyzed and interpreted the data, drafted the manuscript, and revised and approved the final version of the manuscript. Gaburo-Júnior N and Franco LAM performed the experiments and revised and approved the final version of the manuscript. Gouvêa ACRC, Leite LAS and Diz MPE enrolled the patients, collected the clinical data, and revised and approved the final version of the manuscript. Folgueira MAAK conceived the study, analyzed and interpreted the data, drafted the manuscript, and revised and approved the final version of the manuscript.
We acknowledge the helpful assistance of Dr. Rossana Veronica Mendoza Lopez for the statistical analysis. The São Paulo Research Foundation (FAPESP, grant #2012/12306-4) supported this work. Simone Maistro received a postdoctoral scholarship from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, PVE #029/2012), and received a PhD scholarship grant from the São Paulo Research Foundation (FAPESP, #2011/09572-1). Tauana Rodrigues Nagy and Gláucia Fernanda de Lima Pereira received a scholarship grant from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior. Maria Aparecida Azevedo Koike Folgueira received a research grant from the Conselho Nacional de Desenvolvimento Científico e Tecnológico, Brazil (CNPq-308876/2017-2).
Clinical and pathological characteristics of breast cancer patients, BRCA sequencing, and the multiplex ligation-dependent probe amplification (MLPA) results.
| ID | Age Years | HT | HG | ER (%) | PR (%) | HER2 | Ki67 (%) | Molecular Subtype | CS | FH | BRCA | MLPA |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 71 | IDC | 2 | 100 | 80 | Neg. | 25 | Luminal B | III | Yes | wt | wt |
| 2 | 80 | IDC | 2 | 80 | 80 | Neg. | 30 | Luminal B | ND | Yes | BRCA2 | wt |
| 3 | 66 | IDC | 1 | 95 | 80 | Neg. | 15 | Luminal B | II | Yes | wt | ND |
| 4 | 61 | IDC | 2 | 95 | Neg. | Neg. | 20 | Luminal B | II | Yes | wt | wt |
| 5 | 61 | IDC | 2 | 100 | 100 | Neg. | 12 | Luminal A | II | Yes | wt | wt |
| 6 | 74 | IDC | 1 | 100 | 100 | Neg. | 10 | Luminal A | I | Yes | wt | wt |
| 7 | 66 | IDC | 2 | 60 | 66 | Neg. | 30 | Luminal B | I | Yes | wt | wt |
| 8 | 61 | IDC | 2 | Pos. | Pos. | Neg. | 30 | Luminal B | III | Yes | wt | wt |
| 9 | 73 | IDC | 2 | 100 | 100 | Neg. | 10 | Luminal A | I | Yes | wt | wt |
| 10 | 57 | IDC | ND | 90 | 70 | Neg. | 10 | Luminal A | I | Yes | wt | wt |
| 11 | 73 | IDC | 2 | 95 | Neg. | Neg. | 15 | Luminal B | II | Yes | wt | wt |
| 12 | 73 | IDC | 3 | 100 | 5 | Neg. | ND | Luminal | I | Yes | wt | wt |
| 13 | 59 | IDC | 1 | Neg. | Neg. | Pos. | 18 | HER 2 | III | Yes | wt | wt |
| 14 | 62 | IDC | 2 | 90 | 70 | Neg. | ND | Luminal | II | Yes | wt | wt |
| 16 | 60 | IDC | 1 | 90 | 100 | Neg. | 8 | Luminal A | II | Yes | wt | wt |
| 17 | 56 | IDC | 3 | Pos. | Pos. | Neg. | 80 | Luminal B | III | Yes | BRCA1 | wt |
| 18 | 56 | IDC | 3 | Neg. | Neg. | Neg. | 30 | TN | I | Yes | wt | wt |
| 19 | 63 | IDC | 2 | 10 | Neg. | Neg. | 20 | Luminal B | I | Yes | wt | wt |
| 20 | 65 | IDC | 2 | 50 | Neg. | Neg. | 30 | Luminal B | II | Yes | wt | wt |
| 21 | 67 | IDC | 1 | 100 | 100 | Neg. | 30 | Luminal B | II | Yes | wt | wt |
| 22 | 56 | IDC | 2 | 66 | 1 | Neg. | 30 | Luminal B | II | Yes | wt | wt |
| 23 | 62 | IDC | 2 | 95 | 1 | Neg. | 18 | Luminal B | I | Yes | wt | wt |
| 24 | 76 | IDC | 3 | Neg. | Neg. | Pos. | 40 | HER2 | III | Yes | wt | wt |
| 25 | 60 | IDC | 2 | Neg. | Neg. | Neg. | 65 | TN | ND | Yes | wt | wt |
| 26 | 60 | IDC | 2 | Pos. | Neg. | Neg. | 30 | Luminal B | II | Yes | wt | wt |
| 27 | 56 | IDC | 2 | Pos. | Pos. | Neg. | 30 | Luminal B | ND | Yes | wt | wt |
| 28 | 63 | IDC | 2 | 66 | 66 | Neg. | 30 | Luminal B | ND | Yes | wt | wt |
| 29 | 58 | IDC | 2 | Neg. | Neg. | Neg. | 33 | TN | ND | Yes | BRCA1 | wt |
| 30 | 62 | IDC | 1 | 100 | Neg. | Neg. | 5 | Luminal A | III | Yes | wt | wt |
| 31 | 60 | IDC | 2 | 66 | 66 | Neg. | 30 | Luminal B | ND | Yes | wt | wt |
| 32 | 56 | IDC | 1 | 90 | 70 | Neg. | 10 | Luminal A | I | No | wt | wt |
| 33 | 55 | IDC | 1 | Neg. | Neg. | Neg. | ND | TN | I | No | wt | wt |
| 34 | 61 | IDC | 2 | 95 | 15 | Neg. | 20 | Luminal B | II | Yes | wt | wt |
| 35 | 68 | IDC | 2 | 66 | 66 | Neg. | 10 | Luminal A | II | Yes | wt | wt |
| 36 | 63 | IDC | 1 | 90 | 80 | Neg. | 10 | Luminal A | I | Yes | wt | wt |
| 37 | 62 | IDC | 2 | 95 | 0,1 | Pos. | 40 | Luminal B | I | Yes | wt | wt |
| 38 | 59 | IDC | 2 | 40 | 75 | Neg. | 20 | Luminal B | I | Yes | wt | wt |
| 39 | 63 | IDC | 2 | 100 | 100 | Neg. | 13 | Luminal A | II | Yes | wt | wt |
| 40 | 63 | IDC | 2 | 100 | 30 | Pos. | 20 | Luminal B | III | Yes | wt | wt |
| 41 | 62 | IDC | 2 | Pos. | Pos. | Pos. | ND | Luminal | II | Yes | wt | wt |
| 42 | 77 | IDC | 3 | Neg. | Neg. | Pos. | 70 | HER2 | III | Yes | wt | wt |
| 43 | 65 | IDC | 2 | >50 | >50 | Neg. | 5-30 | Luminal | II | Yes | wt | wt |
| 44 | 56 | IDC | 3 | 90 | Neg. | Neg. | 30-40 | Luminal B | II | Yes | BRCA2 | wt |
| 45 | 64 | IDC | 2 | Pos. | Pos. | Neg. | ND | Luminal | II | Yes | wt | wt |
| 46 | 55 | IDC | 2 | Neg. | Neg. | Pos. | 10 | HER2 | III | Yes | wt | wt |
| 47 | 58 | IDC | 3 | Neg. | Neg. | Neg. | 70 | TN | III | Yes | wt | BRCA1 |
| 48 | 75 | IDC | 1 | >66 | >66 | Neg. | <15 | Luminal A | I | Yes | wt | ND |
| 49 | 79 | IDC | 2 | Neg. | Neg. | Pos. | 40 | HER2 | II | Yes | wt | wt |
| 50 | 80 | IDC | 2 | Pos. | Pos. | Neg. | ND | Luminal | II | Yes | wt | wt |
ID: Patient identification; HT: Histological type; HG: Histological grade; ER: Estrogen receptor; PR: Progesterone receptor; CS: Clinical stage; FH: Family history for breast and/or ovarian cancer; ND: Not determined; wt: Wild type; MLPA: Multiplex ligation-dependent probe amplification.
BRCA1 variants.
| Exon | HGVS Nucleotide | HGVS Protein | Protein | Other names | Type | Localization (GRCh37) | NCBI 1000 Genomes Browser | Global MAF dbSNP | Allele Frequency ExAC | Global MAF 1000 genomes | ESP | gnomAD | TOPMed | ABraOM | SIFT | PolyPhen | Provean | Align-GVGD (Pufferfish) | Human Splicing Finder | BRCA Exchange | BRCA Mutation Database | BRCA Share™ | ClinVar | Interpretation | n |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | c.-19-115T>C | − | − | IVS1-115T>C | 5'UTR | 17: 41276247 | rs3765640 | 0.35363 (G) | − | 0,35363 | − | 0,31688 | 0,30248 | 0.304260 | − | − | − | − | Mutant type not implemented in HSF yet | Benign/Little Clinical Significance | ND | ND | Benign | Benign | 9 |
| 2 | c.81-14C>T | − | − | IVS2-14C>T | IVS | 17: 41267810 | rs80358006 | − | − | − | 0.00069 | − | 0,00052 | 0.001642 | − | − | − | − | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign/Likely Benign | Benign/Likely Benign | 1 |
| 3 | c.134+111C>T | − | − | IVS3+111C>T | IVS | 17: 41267632 | rs8176100 | 0.00379 (A) | − | 0,00379 | − | 0,00227 | 0.00128 | − | − | − | − | − | Creation of an intronic ESE site. Probably no impact on splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 1 |
| 6 | c.301+43A>G | − | − | IVS6+43A>G | IVS | 17: 41256841 | − | − | − | − | − | − | − | − | − | − | − | − | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | ND | ND | ND | ND | Uncertain Significance | 1 |
| 7 | c.441+36_441+49delCTTTTCTTTTTTTT | − | − | IVS7+36del14 | IVS | 17: 41256090_41256103 | rs373413425 | − | − | − | − | − | − | 0.295230 | − | − | − | − | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Not Yet Reviewed | ND | 1-Neutral | Benign | Benign | 23 |
| 7 | c.441+36C>T | − | − | IVS7+36C>T | IVS | 17: 41256103 | rs45569832 | − | − | − | − | 0,00009 | − | − | − | − | − | − | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Not Yet Reviewed | ND | 3-UV | Uncertain significance? | Uncertain Significance | 2 |
| 7 | c.441+41C>T | − | − | IVS7+41C>T | IVS | 17: 41256098 | rs45489593 | − | 0,00024 | − | − | 0,00104 | − | − | − | − | − | − | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Not Yet Reviewed | ND | 1-Neutral | Uncertain significance? | Uncertain Significance | 1 |
| 7 | c.442-34C>T | − | − | IVS7-34C>T | IVS | 17: 41251931 | rs799923 | 0.09864 (A) | 0,17379 | 0,09864 | 0,17569 | 0,17303 | 0,14802 | 0.200328 | − | − | − | − | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Not Yet Reviewed | ND | 1-Neutral | Benign | Benign | 15 |
| 9 | c.548-58delT | − | − | c.IVS8-58delT | IVS | 17: 41249364 | rs8176144 | 0.33486 (AAAAAA) | − | − | 0,27833 | 0.3005 | 0,28382 | − | − | − | − | − | Alteration of an intronic ESS site. Probably no impact on splicing. | ND | ND | 1-Neutral | Benign | Benign | 2 |
| 9 | c.591C>T | p.Cys197= | C197= | 710C>T | Syn | 17: 41249263 | rs1799965 | 0.00040 (A) | 0,00147 | 0.00040 | 0,00123 | 0,00178 | 0,00076 | − | − | − | Neutral | − | Activation of an exonic cryptic donor site. Creation of an exonic ESS site. Potential alteration of splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 1 |
| 11 | c.1067A>G | p.Gln356Arg | Q356R | 1186A>G | M | 17: 41246481 | rs1799950 | 0.02177 (C) | 0,04407 | 0,02177 | 0,0459 | 0,05196 | 0,04129 | 0.049261 | Deleterious (0.01) | Probably Damaging (0.988) | Deleterious | Class C0 | Creation of an exonic ESS site. Alteration of an exonic ESE site. Potential alteration of splicing. | Benign/Little Clinical Significance | 1-Not pathogenic or of no clinical significance | 1-Neutral | Benign | Benign | 6 |
| 11 | c.1971A>G | p.Gln657= | Q657= | 2090 A>G | Syn | 17: 41245577 | rs28897679 | 0.00639 (C) | 0,00217 | 0,00639 | 0,00869 | 0,00605 | 0,00741 | 0.005747 | − | − | Neutral | − | Creation of an exonic ESS site. Alteration of an exonic ESE site. Potential alteration of splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 1 |
| 11 | c.2077G>A | p.Asp693Asn | D693N | 2196G>A | M | 17: 41245471 | rs4986850 | 0.03355 (T) | 0.05681 | 0,03355 | 0,05429 | 0.05451 | 0.05336 | 0.056650 | Tolerated (0.08) | Benign (0.01) | Neutral | Class C0 | Alteration of an exonic ESE site. Potential alteration of splicing. | Benign/Little Clinical Significance | 1-Not pathogenic or of no clinical significance | 1-Neutral | Benign | Benign | 9 |
| 11 | c.2082C>T | p.Ser694= | S694= | 2201C>T | Syn | 17: 41245466 | rs1799949 | 0.33646 (A) | 0,34827 | 0,33646 | 0,29568 | 0,31633 | 0,30145 | 0.302956 | − | − | Neutral | − | Activation of an exonic cryptic donor site. Alteration of an exonic ESE site. Potential alteration of splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 22 |
| 11 | c.2311T>C | p.Leu771= | L771L | 2430T>C | Syn | 17: 41245237 | rs16940 | 0.33526 (G) | 0,34196 | 0,33526 | 0,27764 | 0,30018 | 0,28384 | 0.282430 | − | − | Neutral | − | Creation of an exonic ESS site. Potential alteration of splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 22 |
| 11 | c.2596C>T | p.Arg866Cys | R866C | 2715C>T | M | 17: 41244952 | rs41286300 | − | 0,0001 | − | − | 0,00016 | 0,00017 | − | Deleterious (0) | Probably Damaging (1) | Deleterious | Class C65 | Creation of an exonic ESS site. Potential alteration of splicing. | Benign/Little Clinical Significance | 1-Not pathogenic or of no clinical significance | 1-Neutral | Benign | Benign | 1 |
| 11 | c.2612C>T | p.Pro871Leu | P871L | 2731C>T | M | 17: 41244936 | rs799917 | 0.45607 (G) | 0,41005 | 0,54393 | 0,49316 | − | 0,4893 | 0.450739 | Tolerated (1) | Benign (0) | Neutral | Class C0 | Creation of an exonic ESS site. Alteration of an exonic ESE site. Potential alteration of splicing. | Benign/Little Clinical Significance | 1-Not pathogenic or of no clinical significance | 1-Neutral | Benign | Benign | 26 |
| 11 | c.3113A>G | p.Glu1038Gly | E1038G | 3232A>G | M | 17: 41244435 | rs16941 | 0.33566 (C) | 0,34287 | 0,33566 | 0,27903 | 0,30081 | 0,28456 | 0.282430 | Tolerated (0.16) | Possibly Damaging (0.606) | Deleterious | Class C0 | Creation of an exonic ESS site. Alteration of an exonic ESE site. Potential alteration of splicing. | Benign/Little Clinical Significance | 1-Not pathogenic or of no clinical significance | 1-Neutral | Benign | Benign | 22 |
| 11 | c.3119G>A | p.Ser1040Asn | S1040N | 3238G>A | M | 17: 41244429 | rs4986852 | 0.00978 (T) | 0.00978 (T) | 0,00978 | − | 0,01109 | 0,01571 | 0.035304 | Tolerated (0.21) | Possibly Damaging (0.831) | Neutral | Class C0 | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Benign/Little Clinical Significance | 1-Not pathogenic or of no clinical significance | 1-Neutral | Benign | Benign | 5 |
| 11 | c.3305A>G | p.Asn1102Ser | N1102S | − | M | 17: 41244243 | rs80356900 | − | 0,00002 | − | 0,00008 | 0,00001 | 0,00001 | − | Tolerated (0.17) | Benign (0.156) | Deleterious | Class C0 | Creation of an exonic ESS site. Potential alteration of splicing. | Not Yet Reviewed | 2-Likely not pathogenic or of little clinical significance | 3-UV | Uncertain significance? | Uncertain Significance | 1 |
| 11 | c.3548A>G | p.Lys1183Arg | K1183R | 3667A>G | M | 17: 41244000 | rs16942 | 0.35264 (C) | 0,34901 | 0,35264 | 0,29525 | 0,31548 | 0,30133 | 0.299672 | Tolerated (1) | Benign (0) | Neutral | Class C0 | Creation of an exonic ESS site. Alteration of an exonic ESE site. Potential alteration of splicing. | Benign/Little Clinical Significance | 1-Not pathogenic or of no clinical significance | 1-Neutral | Benign | Benign | 22 |
| 11 | c.3752G>A | p.Cys1251Tyr | C1251Y | − | M | 17: 41243796 | rs879254079 | − | − | − | − | − | − | − | Tolerated (1) | Benign (0.001) | Neutral | Class C0 | Alteration of an exonic ESE site. Potential alteration of splicing. | Not Yet Reviewed | ND | ND | Uncertain significance? | Uncertain Significance | 1 |
| 11 | c.4039A>G | p.Arg1347Gly | R1347G | 4158A>G | M | 17: 41243509 | rs28897689 | 0.00060 (C) | 0.00398 | 0,0006 | 0,00484 | 0,00423 | 0,00481 | 0.005747 | Tolerated (0.09) | Benign (0.071) | Neutral | Class C0 | Creation of an exonic ESS site. Alteration of an exonic ESE site. Potential alteration of splicing. | Benign/Little Clinical Significance | 1-Not pathogenic or of no clinical significance | 1-Neutral | Benign | Benign | 2 |
| 12 | c.4113G>A | p.Gly1371= | G1371= | − | Syn | 17: 41243033 | rs147448807 | 0.00160 (T) | 0.00050 | 0,0016 | 0,00123 | 0,00156 | − | − | − | − | Neutral | − | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Likely benign | ND | 2-Likely Neutral | Likely Benign | Likely Benign | 1 |
| 13 | c.4308T>C | p.Ser1436= | S1436= | 4427T>C | Syn | 17: 41234470 | rs1060915 | 0.33626 (G) | 0,3431 | 0,33626 | 0,27956 | 0,30142 | 0,28489 | 0.283251 | − | − | Neutral | − | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 22 |
| 15 | c.4485-63C>G | − | − | IVS 14-63C>G | IVS | 17: 41226601 | rs273900734 | 0.35344 (C) | − | 0,35344 | − | 0,31645 | 0,30211 | 0.300493 | − | − | − | − | − | Benign/Little Clinical Significance | ND | ND | Benign | Benign | 2 |
| 16 | c.4837A>G | p.Ser1613Gly | S1613G | 4956A>G | M | 17: 41223094 | rs1799966 | 0.35583 (C) | − | 0,35583 | 0,29817 | − | 0,30333 | 0.300987 | Tolerated (0.11) | Benign (0.038) | Neutral | Class C0 | Alteration of an exonic ESE site. Potential alteration of splicing. | Benign/Little Clinical Significance | 1-Not pathogenic or of no clinical significance | 1-Neutral | Benign | Benign | 22 |
| 16 | c.4987-92A>G | − | − | IVS16-92A>G | IVS | 17: 41219804 | rs8176233 | 0.35463 (C) | − | 0,35463 | − | 0,31276 | 0,30294 | 0.300493 | − | − | − | − | Alteration of an exonic ESE site. Potential alteration of splicing.- | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 3 |
| 16 | c.4987-68A>G | − | − | VS16-68A>G | IVS | 17: 41219780 | rs8176234 | 0.35463 (C) | − | 0,35463 | − | 0,31483 | 0,30295 | 0.301314 | − | − | − | − | Alteration of an exonic ESE site. Potential alteration of splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 3 |
| 17 | c.5074+2T>C | − | − | IVS17+2T>C | SS | 17: 41219623 | rs80358089 | − | − | − | − | − | − | − | − | − | − | − | Alteration of the WT donor site, most probably affecting splicing | Pathogenic | 5-Definitely pathogenic | ND | Pathogenic? | Pathogenic | 1 |
| 17 | c.5075-53C>T | − | − | IVS17-53C>T | IVS | 17: 41216021 | rs8176258 | 0.01098 (A) | − | 0,01098 | 0,01708 | 0,01825 | 0,01721 | 0.018062 | − | − | − | − | Alteration of an intronic ESS site. Probably no impact on splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 2 |
| 18 | c.5123C>A | p.Ala1708Glu | A1708E | 5242C>A | M | 17: 41215920 | rs28897696 | − | 0,02487 | − | 0.00023 (T) | − | − | − | Deleterious (0) | Possibly Damaging (0.633) | Neutral | Class C65 | Activation of an exonic cryptic acceptor site, with presence of one or more cryptic branch point(s). Creation of an exonic ESS site. Alteration of an exonic ESE site. Potential alteration of splicing. | Pathogenic | 5-Definitely pathogenic | 5-Causal | Pathogenic? | Pathogenic? | 1 |
| 18 | c.5152+66G>A | − | − | IVS18+66G>A | IVS | 17: 41215825 | rs3092994 | 0.34245 (T) | − | 0,34245 | − | 0,31394 | 0,29599 | 0.291461 | − | − | − | − | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 3 |
| 21 | c.5304C>T | p.Cys1768= | C1769C | − | M | 17: 41203108 | rs138493864 | 0.00060 (A) | 0.00002 | 0,0006 | 0,00015 | 0,00013 | 0,00009 | − | − | − | Neutral | − | Creation of an exonic ESS site. Potential alteration of splicing. | Likely benign | ND | ND | Likely Benign | Likely Benign | 1 |
HGVS: Human Genome Variation Society; MAF: Minor allele frequency; ESP: NHLBI Exome Sequencing Project Exome Variant Server; gnomAD: The Genome Aggregation Database; TOPMed: Trans-Omics for Precision Medicine; ABraOM: Brazilian genomic variants; SIFT: Sorting intolerant from tolerant; PolyPhen: Polymorphism Phenotyping; Provean: Protein Variation Effect Analyzer; Align-GVGD: Class C0 (less probable to interfere with protein function), C15, C25, C35, C45, C55, C65 (more probable to interfere with protein function); Syn: Synonymous; IVS: Intervening sequence; M: Missense; SS: splice site; ND, Not determined; n: Number of patients harboring the variant
BRCA2 variants.
| Exon | HGVS Nucleotide | HGVS Protein | Protein Abbrev | Other names | Type | Localization (GRCh37) | NCBI 1000 Genomes Browser | Global MAF dbSNP | Allele Frequency ExAC | Global MAF 1000 genomes | ESP | gnomAD | TOPMed | ABraOM | SIFT | PolyPhen | Provean | Align-GVGD (Pufferfish) | Human Splicing Finder | BRCA Exchange | BRCA Mutation Database | BRCA Share™ | ClinVar | Interpretation | n |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | c.-26G>A | − | − | 203G>A | 5'UTR | 13: 32890572 | rs1799943 | 0.20927 (A) | 0,24652 | 0,20927 | 0,20883 | 0,22032 | 0,21567 | 0.217570 | − | − | − | − | ND | Benign/Little Clinical Significance | ND | ND | Benign | Benign | 21 |
| 2 | c.-15A>C | − | − | 214A>C | 5'UTR | 13: 32890583 | rs138705202 | 0.00080 (C) | 0.00022 | 0,0008 | 0,00038 | 0,00064 | 0,00076 | 0.002463 | − | − | − | − | ND | Not Yet Reviewed | ND | ND | Benign/Likely Benign | Likely benign | 1 |
| 2 | c.-11C>T | − | − | 218C>T | 5'UTR | 13: 32890587 | rs76874770 | 0.00439 (T) | 0,00163 | 0,00439 | 0,00584 | 0,0051 | 0,00546 | 0.007389 | − | − | − | − | ND | Benign/Little Clinical Significance | ND | ND | Benign | Benign | 2 |
| 2 | c.2T>G | p.Met1Arg | M1R | − | M | 13: 32890599 | rs80358547 | − | 0,00001 | − | − | 0,00001 | − | − | Damaging (0.00) | Probably Damaging (0.998) | Deleterious | Class C65 | ND | Not Yet Reviewed | 5-Definitely pathogenic | 5-Causal | Pathogenic? | Pathogenic | 1 |
| 3 | c.125A>G | p.Tyr42Cys | Y42C | 353A>G | M | 13: 32893271 | rs4987046 | 0.00080 (G) | 0,0017 | 0,0008 | 0,00246 | 0,00162 | 0,00158 | 0.001642 | Tolerate (0.12) | Benign (0.090) | Neutral | Class C0 | Activation of an exonic cryptic donor site. Potential alteration of splicing. | Benign/Little Clinical Significance | 1-Not pathogenic or of no clinical significance | 1-Neutral | Benign | Benign | 1 |
| 4 | c.425+33A>G | − | − | IVS4+33A>C | IVS | 13: 32899354 | rs200065709 | 0.00060 (G) | 0,00052 | 0,0006 | 0,00031 | 0.00010 | 0,00029 | 0.000821 | − | − | − | − | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Not Yet Reviewed | ND | 2-Likely Neutral | Benign/Likely Benign | Likely benign | 1 |
| 4 | c.425+67A>C | − | − | IVS4+67A>C | IVS | 13: 32899388 | rs11571610 | 0.07428 (C) | − | 0,07428 | − | 0,03064 | 0,03973 | 0.045156 | − | − | − | − | Alteration of an intronic ESS site. Probably no impact on splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 6 |
| 6 | c.517-19C>T | − | − | IVS6-19C>T | IVS | 13: 32900617 | rs11571623 | 0.00819 (T) | 0,00219 | 0,00819 | 0,00738 | 0,00586 | 0,007 | 0.003284 | − | − | − | − | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 2 |
| 8 | c.681+56C>T | − | − | IVS8+56C>T | IVS | 13: 32903685 | rs2126042 | 0.18590 (T) | − | 0,1859 | − | 0,21627 | 0,20076 | 0.184729 | − | − | − | − | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Not Yet Reviewed | ND | 1-Neutral | Benign | Benign | 17 |
| 10 | c.865A>C | p.Asn289His | N289H | 1093A>C | M | 13: 32906480 | rs766173 | 0.07368 (C) | − | 0,07368 | 0,03055 | 0,03055 | 0,03968 | 0.045156 | Damaging (0.003) | Benign (0.278) | Neutral | Class C0 | Alteration of an exonic ESE site. Potential alteration of splicing | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 7 |
| 10 | c.1114A>C | p.His372Asn | H372N | 1342 A>C | M | 13: 32906729 | rs144848 | 0.24940 (C) | 0,27793 | 0,2494 | − | 0,22303 | 0,23657 | 0.259442 | Tolerated (0.35) | Benign (0.00) | Neutral | Class C0 | Alteration of an exonic ESE site. | Benign/Little Clinical Significance | 1-Not pathogenic or of no clinical significance | 1-Neutral | Benign | Benign | 19 |
| 10 | c.1365A>G | p.Ser455= | S455= | 1593A>G | Syn | 13: 32906980 | rs1801439 | 0.07368 (G) | 0,05178 | 7368 | 0,03101 | 0,03048 | 0,03968 | 0.045156 | − | − | Neutral | − | Alteration of an exonic ESE site. Potential alteration of splicing | ND | ND | 1-Neutral | Benign | Benign | 7 |
| 10 | c.1514T>C | p.Ile505Thr | I505T | M | 13: 32907129 | rs28897708 | 0.00040 (C) | 0,00072 | 0,0004 | 0,00077 | 0,00083 | 0,00065 | 0.000821 | Tolerated (0.1) | Possibly Damaging (0.651) | Neutral | Class C0 | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Benign/Little Clinical Significance | 1-Not pathogenic or of no clinical significance | 1-Neutral | Benign | Benign | 1 | |
| 10 | c.1909+92_1909+96del | − | − | IVS10+92del5 | IVS | 13: 32907615-32907620 | rs144549870 | 0.01577 (TAT) | − | − | − | − | − | 0.006568 | − | − | − | − | − | ND | ND | ND | Benign | Benign | 2 |
| 10 | c.1910-74T>C | − | − | IVS10-74T>C | IVS | 13: 32910328 | rs2320236 | 0.17452 (C) | − | 0,17452 | − | 0,20561 | 0,20561 | rs2320236 | − | − | − | − | Creation of an intronic ESE site. Probably no impact on splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 14 |
| 10 | c.1910-51G>T | − | − | IVS10-51G>T | IVS | 13: 32910351 | rs11571651 | 0.07348 (T) | 0,04934 | 0,07348 | 0,03056 | 0,03041 | 0,03968 | 0.045977 | − | − | − | − | Alteration of an intronic ESS site. Probably no impact on splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 6 |
| 11 | c.2229T>C | p.His743= | H743= | 2457T>C | Syn | 13: 32910721 | rs1801499 | 0.07348 (C) | 0,05158 | 0.07348 | 0,03129 | 0,03065 | 0,03972 | 0.045156 | − | − | Neutral | − | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 7 |
| 11 | c.2350A>G | p.Met784Val | M784V | 2578A>G | M | 13: 32910842 | rs11571653 | 0.00359 (G) | 0,00031 | 0,00359 | − | 0,00023 | 0,00022 | 0.002463 | Tolerated (1.00) | Benign (0.00) | Neutral | Class C0 | Creation of an exonic ESS site. Potential alteration of splicing. | Benign/Little Clinical Significance | 3-Uncertain | 3-UV | Benign | Benign | 1 |
| 11 | c.2971A>G | p.Asn991Asp | N991D | 3199A>G | M | 13: 32911463 | rs1799944 | 0.08007 (G) | 0,05341 | 0,08007 | 0,03725 | 0,03723 | 0,0461 | 0.046798 | Tolerated (1.00) | Benign (0.00) | Neutral | Class C0 | Alteration of an exonic ESE site. Potential alteration of splicing | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 7 |
| 11 | c.3264T>C | p.Pro1088= | P1088= | 3492T>C | Syn | 13: 32911756 | rs36060526 | 0.00679 (C) | 0.00238 | 0,00679 | 0,00756 | 0,00762 | 0,00756 | 0.006568 | − | − | Neutral | − | ND | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 1 |
| 11 | c.3371A>G | p.Gln1124Arg | Q1124R | M | 13: 32911863 | rs1555283204 | − | − | − | − | − | − | − | Damaging (0.01) | Probably Damaging (1.00) | Deleterious | Class C35 | Activation of an exonic cryptic donor site. Potential alteration of splicing. | Not Yet Reviewed | ND | ND | Uncertain significance | Uncertain significance | 1 | |
| 11 | c.3396A>G | p.Lys1132= | L1132= | 3624A>G | Syn | 13: 32911888 | rs1801406 | 0.26677 (G) | 0,29449 | 0,26677 | 0,27984 | 0,29762 | 0,28221 | 0.283251 | − | − | Neutral | − | Alteration of an exonic ESE site. Potential alteration of splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 23 |
| 11 | c.3807T>C | p.Val1269= | V1269= | 4035T>C | Syn | 13: 32912299 | rs543304 | 0.16813 (C) | 0,18985 | 0,16813 | 0,19111 | 0,18144 | 0,18622 | 0.187192 | − | − | Neutral | − | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 23 |
| 11 | c.4068G>A | p.Leu1356= | L1356= | 4296G>A | Syn | 13: 32912560 | rs28897724 | 0.00040 (A) | 0,00305 | 0,0004 | 0,00315 | 0.00245 | 0,00312 | 0.002463 | − | − | Neutral | − | ND | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 1 |
| 11 | c.4090A>C | p.Ile1364Leu | I1364L | 4318A>C | M | 13: 32912582 | rs56248502 | 0.00439 (C) | 0,00172 | 0,00439 | 0,00631 | 0,00577 | 0.006568 | Tolerated (0.76) | Benign (0.001) | Neutral | Class C0 | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 2 | |
| 11 | c.4258G>T | p.Asp1420Tyr | D1420Y | 4486G>T | M | 13: 32912750 | rs28897727 | 0.00399 (T) | 0,0068 | 0,00399 | 0,00396 | 0,00794 | 0,00425 | 0.001642 | Damaging (0.01) | Benign (0.030) | Deleterious | Class C15 | ND | Benign/Little Clinical Significance | 1-Not pathogenic or of no clinical significance | 1-Neutral | Benign | Benign | 1 |
| 11 | c.5418A>G | p.Glu1806= | E1806= | 5646A>G | Syn | 13: 32913910 | rs34351119 | 0.00679 (G) | 0,00233 | 0,00679 | 0,0083 | 0,00764 | 0,00785 | 0.006568 | − | − | Neutral | − | ND | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 1 |
| 11 | c.5640T>G | p.Asn1880Lys | N1880K | 5868T>G | M | 13: 32914132 | rs11571657 | 0.00220 (G) | 0,00076 | 0,0022 | 0,00315 | 0,00264 | 0,00294 | 0.000821 | Damaging (0.05) | Benign (0.167) | Neutral | Class C0 | Creation of an exonic ESS site. Potential alteration of splicing. | Benign/Little Clinical Significance | 2-Likely not pathogenic or of little clinical significance | 2-Likely Neutral | Benign/Likely Benign | Likely benign | 1 |
| 11 | c.5645C>A | p.Ser1882Ter | S1882X | 5873C>A | N | 13: 32914137 | rs80358785 | − | 0,00002 | − | − | 0,00002 | 0,00002 | − | − | − | − | − | Alteration of an exonic ESE site. Potential alteration of splicing. | Pathogenic | 5-Definitely pathogenic | 5-Causal | Pathogenic | Pathogenic | 1 |
| 11 | c.5744C>T | p.Thr1915Met | T1915M | 5972C>T | M | 13: 32914236 | rs4987117 | 0.00859 (T) | 0,02114 | 0,02114 | 0,02114 | 0.00859 (T) | 0,01744 | 0.017241 | Tolerated (0.13) | Benign (0.000) | Neutral | Class C0 | Creation of an exonic ESS site. Potential alteration of splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 2 |
| 11 | c.5768A>C | p.Asp1923Ala | D1923A | 5996A>C | M | 13: 32914260 | rs45491005 | 0.00020 (C) | − | 0,0002 | 0,0002 | 0.00054 | 0.00105 | − | Tolerated (0.29) | Benign (0.144) | Deleterious | Class C0 | Alteration of an exonic ESE site. Potential alteration of splicing. | Benign/Little Clinical Significance | 2-Likely not pathogenic or of little clinical significance | 2-Likely Neutral | Benign | Likely benign | 1 |
| 11 | c.6841+53delTATTCAGTAG | − | − | − | IVS | 13: 32915384-32915394 | − | − | − | − | − | − | − | − | − | − | − | − | Alteration of an intronic ESS site. Probably no impact on splicing. | ND | ND | ND | ND | Uncertain Significance | 1 |
| 11 | c.6841+80delTTAA | − | − | IVS11+80delTTAA | IVS | 13: 32915411-32915414 | rs11571661 | 0.26578 (AA) | − | − | − | − | − | 0.279605 | − | − | − | − | Creation of an intronic ESE site. Probably no impact on splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 7 |
| 14 | c.7017G>C | p.Lys2339Asn | K2339N | 7245 G>C | M | 13: 32929007 | rs45574331 | 0.00679 (C) | 0,00228 | 0,00679 | 0,00808 | 0,00764 | 0,00786 | 0.006568 | Damaging (0.01) | Benign (0.105) | Neutral | Class C0 | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Benign/Little Clinical Significance | ND | 2-Likely Neutral | Benign | Benign | 1 |
| 14 | c.7242A>G | p.Ser2414= | S2114= | 7470A>G | Syn | 13: 32929232 | rs1799955 | 0.23263 (G) | − | 0,23263 | 0,21136 | − | 0,22464 | 0.238095 | − | − | Neutral | − | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 19 |
| 14 | c.7319A>G | p.His2440Arg | H2440R | 7547A>G | M | 13: 32929309 | rs4986860 | 0.01038 (G) | 0,00304 | 0,01038 | 0,01054 | 0,00946 | 0,00967 | 0.007389 | Tolerated (0.55) | Benign (0.002) | Neutral | Class C0 | ND | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 1 |
| 14 | c.7397T>C | p.Ala2466Val | A2466V | − | M | 13: 32929387 | rs169547 | 0.02416 (T) | 0,99372 | 0.97584 | 0,9777 | 0,97881 | 0,98191 | 0.983580 | Tolerated (0.98) | Possibly Damaging (0.793) | Neutral | Class C0 | ND | ND | ND | 1-Neutral | Benign | Benign | 50 |
| 14 | c.7435+53C>T | − | − | IVS14+53C>T | IVS | 13: 32929478 | rs11147489 | 0.07248 (T) | − | 0,07248 | − | 0,0301 | 0,03924 | − | − | − | − | − | Creation of an intronic ESE site. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 1 |
| 15 | c.7469T>C | p.Ile2490Thr | I2490T | 7697T>C | M | 13: 32930598 | rs11571707 | 0.01597 (C) | 0,01436 | 0.01597 | 0,00161 | 0,0035 | 0,00913 | 0.021346 | Tolerated (1.00) | Benign (0.010) | Neutral | Class C45 | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 1 |
| 17 | c.7806-14T>C | − | − | IVS16-14T>C | IVS | 13: 32936646 | rs9534262 | 0.46845 (T) | 0,52083 | 0,53155 | 0,52015 | 0,54679 | 0,53151 | 0.523810 | − | − | − | − | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Benign/Little Clinical Significance | ND | 3-UV | Benign | Likely benign | 36 |
| 19 | c.8460A>C | p.Val2820= | V2820= | 8688A>C | Syn | 13: 32944667 | rs9590940 | 0.01438 (C) | 0,00368 | 0,01438 | 0,01299 | 0,01105 | 0,01219 | 0.006568 | − | − | Neutral | − | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 2 |
| 19 | c.8487+47C>T | − | − | IVS19+47C>T | IVS | 13: 32944741 | rs11571744 | 0.01617 (T) | − | 0,01617 | 0,01523 | − | 0,01481 | 0.006568 | − | − | − | − | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Benign/Little Clinical Significance | ND | 3-UV | Benign | Benign | 3 |
| 20 | c.8632+132dup | − | − | c.IVS20+132insC | IVS | 13: 32945368-32945369 | rs201392123 | 0.00899 (CC) | − | 0.00899 | − | 0,00619 | 0,00754 | 0.002627 | − | − | − | − | ND | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 2 |
| 21 | c.8755-66T>C | − | − | IVS21-66T>C | IVS | 13: 32953388 | rs4942486 | 0.48842 (T) | − | 0,51158 | − | 0,52569 | 0,51037 | 0.508210 | − | − | − | − | Alteration of an intronic ESS site. Probably no impact on splicing. Creation of an intronic ESE site. Probably no impact on splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 38 |
| 22 | c.8851G>A | p.Ala2951Thr | A2951T | 9079G>A | M | 13: 32953550 | rs11571769 | 0.00998 (A) | 0,00785 | 0.00998 | 0,00438 | 0,00363 | 0,00721 | 0.013136 | Damaging (0.00) | Probably Damaging (1.00) | Neutral | Class C55 | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 1 |
| 22 | c.8942A>G | p.Glu2981Gly | E2981G | 9170A>G | M | 13: 32953641 | rs398122716 | − | 0,00001 | − | − | 0,00002 | 0,00001 | − | Tolerated (0.16) | Benign (0.030) | Neutral | Class C65 | ND | ND | ND | 3-UV | Conflicting interpretations of pathogenicity? Likely benign(1);Uncertain significance(3) | Uncertain significance | 1 |
| 23 | c.9038C>T | p.Thr3013Ile | T3013I | − | M | 13: 32953971 | rs28897755 | − | 0,00023 | − | 0,00046 | 0,00019 | 0,0002 | − | Tolerated (0.24) | Probably Damaging (0.875) | Neutral | Class C0 | ND | Benign/Little Clinical Significance | 1-Not pathogenic or of no clinical significance | 1-Neutral | Benign | Benign | 1 |
| 24 | c.9257-83G>A | − | − | IVS24-83G>A | IVS | 13: 32968743 | rs9595456 | 0.05052 (A) | − | 0,05052 | − | 0,04116 | 0.04575 | 0.022989 | − | − | − | − | Creation of an intronic ESE site. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 4 |
| 24 | c.9257-16T>C | − | − | IVS24-16T>C | IVS | 13: 32968810 | rs11571818 | 0.00439 (C) | 0,00439 | 0,00765 | 0,00592 | 0,00548 | 0,00548 | 0.004926 | − | − | − | − | No significant splicing motif alteration detected. This mutation has probably no impact on splicing. | ND | ND | 3-UV | Benign | Likely benign | 1 |
| 27 | c.9730G>A | p.Val3244Ile | V3244I | 9958 G>A | M | 13: 32972380 | rs11571831 | 0.00679 (A) | − | − | 0,0083 | 0,00767 | 0,00787 | − | Tolerated (0.49) | Benign (0.000) | Neutral | Class C0 | ND | Benign/Little Clinical Significance | ND | 2-Likely Neutral | Benign | Benign | 1 |
| 27 | c.9976A>T | p.Lys3326Ter | K3326X | 10204A>T | N | 13: 32972626 | rs11571833 | 0.00439 (T) | 0,00702 | 0,00439 | 0.00646 | 0,00544 | 0,00547 | 0.004926 | − | − | − | − | Creation of an exonic ESS site. Potential alteration of splicing. Alteration of an exonic ESE site. Potential alteration of splicing. | Benign/Little Clinical Significance | 2-Likely not pathogenic or of little clinical significance | 1-Neutral | Benign | Benign | 1 |
| 27 | c.10110G>A | p.Arg3370= | R3370= | − | Syn | 13: 32972760 | rs28897762 | 0.00080 (A) | 0,00147 | 0,0008 | 0,00215 | 0,0014 | 0,00131 | 0.000821 | − | − | Neutral | − | Alteration of an exonic ESE site. Potential alteration of splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 1 |
| 27 | c.10234A>G | p.Ile3412Val | I3412V | 10462 A>G | M | 13: 32972884 | rs1801426 | 0.04493 (G) | 0,02266 | 0,04493 | 0,03729 | 0,0369 | 0,04054 | 0.021346 | Tolerrated (0.34) | Benign (0.002) | Neutral | Class C0 | Alteration of an exonic ESE site. Potential alteration of splicing. | Benign/Little Clinical Significance | ND | 1-Neutral | Benign | Benign | 4 |
HGVS: Human Genome Variation Society; MAF: Minor allele frequency; EXAC: Exome Aggregation Consortium; ESP: NHLBI Exome Sequencing Project Exome Variant Server; gnomAD: The Genome Aggregation Database; TOPMed: Trans-Omics for Precision Medicine; ABraOM: Brazilian genomic variants; SIFT: Sorting Intolerant From Tolerant; PolyPhen: Polymorphism Phenotyping; Provean: Protein Variation Effect Analyzer; Align-GVGD: Class C0 (less probable to interfere with protein function), C15, C25, C35, C45, C55, C65 (more probable to interfere with protein function); Syn: Synonymous; IVS: Intervening sequence; M: Missense; SS: Splice site; ND: Not determined; n: Number of patients bearing the variant
In silico analysis of the alterations in exons 9 and 20 of PIK3CA in postmenopausal patients with breast cancer.
| Sample ID | Age at diagnosis | Molecular Subtype | Exon | Cdna | Protein | Protein | Mutation Type | ID COSMIC | Polyphen | SIFT | Provean | Align-GVGD |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 71 | Luminal B | 9 | c.1634A>C | p.Glu545Ala | E545A | M | COSM12458 | Probably Damaging | Damaging | Deleterious | Class C65 |
| 3 | 66 | Luminal B | 9 | c.1639G>C | p.Glu547Gln | E547Q | M | − | Probably Damaging | Damaging | Neutral | Class C25 |
| 8 | 61 | Luminal B | 20 | c.3075C>T | p.Thr1025= | T1025T | Syn | COSM21451 | − | Tolerated | Neutral | − |
| c.3140A>T | p.His1047Leu | H1047L | M | COSM776 | Benign | Damaging | Neutral | Class C65 | ||||
| 9 | 73 | Luminal A | 9 | c.1629C>T | p.Ile543= | I543I | Syn | COSM5020257 | − | Tolerated | Neutral | − |
| 10 | 57 | Luminal A | 9 | c.1549C>T | p.Leu517= | L517L | Syn | − | − | Tolerated | Neutral | − |
| 17* | 56 | Luminal B | 9 | c.1634A>C | p.Glu545Ala | E545A | M | COSM12458 | Probably Damaging | Damaging | Deleterious | Class C65 |
| 21 | 67 | Luminal B | 9 | c.1550T>C | p.Leu517Pro | L517P | M | − | Benign | Damaging | Neutral | Class C65 |
| 23 | 62 | Luminal B | 20 | c.3140A>T | p.His1047Leu | H1047L | M | COSM776 | Benign | Damaging | Neutral | Class C65 |
| 26 | 60 | Luminal B | 9 | c.1547G>A | p.Arg516Lys | R516K | M | COSM3724545 | Benign | Tolerated | Neutral | Class C25 |
| 20 | c.3170G>A | p.Trp1057* | W1057X | N | COSM6475611 | − | − | − | − | |||
| 32 | 56 | Luminal A | 20 | c.3098A>G | Gln1033Arg | Q1033R | M | COSM303947 | Possible Damaging | Damaging | Neutral | Class C35 |
| 36 | 63 | Luminal A | 9 | c.1634A>C | p.Glu545Ala | E545A | M | COSM12458 | Probably Damaging | Damaging | Deleterious | Class C65 |
| c.1658_1659delGTinsC | p.Ser553Thrfs*7 | S553fs | F | − | − | − | − | − | ||||
| 39 | 63 | Luminal A | 9 | c.1638G>A | p.Gln546= | Q546Q | Syn | COSM5622324 | − | Toleratd | Neutral | − |
| c.1664+46G>A | − | − | IVS | − | − | − | − | − | ||||
| 20 | c.3102G>A | p.Glu1034= | E1034E | Syn | − | − | Tolerated | Neutral | − | |||
| 46 | 55 | HER2 | 9 | c.1634A>C | p.Glu545Ala | E545A | M | COSM12458 | Probably Damaging | Damaging | Deleterious | Class C65 |
| c.1651C>T | p.Leu551= | L551L | Syn | COSM308546 | − | Tolerated | Neutral | − | ||||
| c.1658_1659delGTinsC | p.Ser553Thrfs*7 | S553fs | F | − | − | − | − | − | ||||
| 47* | 58 | TN | 9 | c.1622C>T | p.Ser541Phe | S541F | M | COSM6438100 | Possible Damaging | Damaging | Deleterious | Class C65 |
| 20 | c.3110A>T | p.Glu1037Val | E1037V | M | − | Benign | Damaging | Deleterious | Class C65 |
HGVS: Human Genome Variation Society; SIFT: Sorting Intolerant From Tolerant; PolyPhen: Polymorphism Phenotyping; Provean: Protein Variation Effect Analyzer; Align-GVGD: Class C0 (less probable to interfere with protein function), C15, C25, C35, C45, C55, C65 (more probable to interfere with protein function); Syn: Synonymous; IVS: Intervening Sequence; M: Missense; N: Nonsense. *Patients also harboring pathogenic germline mutations in BRCA1.
In silico analysis of the alterations in exons 9 and 20 of PIK3CAin young patients with breast cancer.
| Sample ID | Age at diagnosis | Molecular Subtype | Exon | cDNA | Protein | p.Asn1044Asp | Mutation Type | ID COSMIC | Polyphen | SIFT | Provean | Align-GVGD |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 452 | 34 | Luminal B | 20 | c.3130A>G | p.Asn1044Asp | N1044D | M | COSM27134 | Probably Damaging | Tolerated | Neutral | Class C15 |
| 454 | 34 | Luminal | 20 | c.3146G>A | p.Gly1049Asp | G1049D | M | COSM308548 | Probably Damaging | Tolerated | Neutral | Class C65 |
| 455 | 28 | Luminal B | 9 | c.1558G>A | p.Asp520Asn | D520N | M | COSM29096 | Benign | Tolerated | Neutral | Class C15 |
| 457 | 33 | Luminal B | 9 | c.1615C>T | p.Pro539Ser | P539S | M | COSM249880 | Probably Damaging | Tolerated | Deleterious | Class C65 |
| c.1664G>A | p.Arg555Lys | R555K | M | COSM1716158 | Probably Damaging | Damaging | Deleterious | Class C25 | ||||
| 468 | 33 | Luminal A | 9 | c.1656G>A | p.Trp552* | W552X | N | COSM37025 | − | − | − | − |
| 477 | 27 | TN | 9 | c.1634A>C | p.Glu545Ala | E545A | M | COSM12458 | Probably Damaging | Damaging | Deleterious | Class C65 |
| 478 | 29 | HER2 | 20 | c.3165G>A | p.Met1055Ile | M1055I | M | COSM9146166 | Benign | Tolerated | Neutral | Class C0 |
| c.3201G>A | p. Leu1067= | L1067L | Syn | − | − | Tolerated | Neutral | − | ||||
| 480 | 31 | TN | 9 | c.1634A>C | p.Glu545Ala | E545A | M | COSM12458 | Probably Damaging | Damaging | Deleterious | Class C65 |
| 20 | c.3203A>C | p.Asn1068Thr | N1068T | M | − | Probably Damaging | Damaging | Neutral | Class C55 | |||
| 483 | 35 | Luminal B | 9 | c.1634A>C | p.Glu545Ala | E545A | M | COSM12458 | Probably Damaging | Damaging | Deleterious | Class C65 |
| 484 | 35 | Luminal B | 9 | c.1593C>A | p.Leu531= | L531L | Syn | − | − | Tolerated | Neutral | − |
| 503 | 35 | HER2 | 20 | c.3150C>T | p.Gly1050= | G1050G | Syn | − | − | Tolerated | Neutral | − |
| 518 | 31 | Luminal B | 9 | c.1615C>T | p.Pro539Ser | P539S | M | COSM249880 | Probably Damaging | Tolerated | Deleterious | Class C65 |
HGVS: Human Genome Variation Society; SIFT: Sorting Intolerant From Tolerant; PolyPhen: Polymorphism Phenotyping; Provean: Protein Variation Effect Analyzer; Align-GVGD: Class C0 (less probable to interfere with protein function), C15, C25, C35, C45, C55, C65 (more probable to interfere with protein function); Syn: Synonymous; M: Missense; N: Nonsense.
BRCA1 and BRCA2 were analyzed for mutations using the Ion AmpliSeq™ BRCA1 and BRCA2 panels (Thermo Fisher Scientific). This panel consists of three primer pools (167 amplicons) covering the entire coding region, including 10-20 bp of non-coding sequences flanking the 5' and 3' ends of each exon. Library preparation was performed using the Ion AmpliSeq™ Library Kit 2.0 and Ion Xpress™ Barcode Adapter 1-96 kit. DNA amplification was performed using 30 ng of DNA with three primer pools and 5x Ion AmpliSeq™ HiFi Master Mix. The PCR cycle included the following: 2 min at 99°C, followed by 19 cycles of 99°C for 15s and 60°C for 4 min, ending with a hold step at 10°C on a Veriti Thermal Cycler (Thermo Fisher Scientific). Next, the three PCR amplicons were mixed (30 µL), and 20 µL was treated with 2 µL FuPa Reagent to partially digest the primer sequences and phosphorylate the amplicons at 50°C for 10 min, followed by 55°C for 10 min, then 60°C for 20 min, and then held at 10°C. Next, sequencing adaptors (A: conjugated to biotin and P1) and barcodes (consisting of short stretches of index sequences that enable sample multiplexing) were ligated to the amplicons using the Ion Xpress™ Barcode Adapters kit (Thermo Fisher Scientific) for 30 min at 22°C, 5 min at 68°C, and 5 min at 72°C, ending with a hold at 10°C. The adaptor-ligated amplicon (libraries) were purified with 45 μL of the Agencourt® AMPure® XP Reagents (Beckman Coulter) and incubated for 5 min at room temperature(22-25°C). The tube was placed in a magnetic rack such that it was incubated for 2 min or until the solution became clear. After the supernatant was removed carefully and discarded without disturbing the pellet, freshly prepared 70% ethanol (150 μL) was added, and the tube was moved side‐to‐side of the magnet to wash the beads, and then the supernatant was discarded (two rounds of purification were repeated). The tube was placed on the magnet, and the beads were air-dried at room temperature for 5 min. The library was subjected to a second round of amplification using 50 μL of Platinum® PCR SuperMix HiFi and 2 μL of Equalizer™ Primers (added to each bead-pellet); the PCR cycles included 98°C for 2 min, followed by 9 cycles of 98°C for 15s and 64°C for 1 min, ending with a hold at 10°C. Then, 10 μL of Equalizer™ Capture was added to each amplified library, mixed by pipetting, and incubated at room temperature for 5 min. Next, 6 μL of washed Equalizer™ beads was added to each tube containing the captured library, mixed, and incubated at room temperature for 5 min. The tube was then placed in the magnet and incubated for 2 min or until the solution became clear. After the supernatant was removed carefully without disturbing the pellet, the Equalizer™ Wash Buffer (150 μL) was added to each reaction, to wash the beads, the tube was moved side‐to‐side of the magnet, and then the supernatant was removed and discarded (two rounds of purification were repeated). Next, the tube was removed from the magnet and 100 μL of Equalizer™ Elution Buffer was added to each pellet, mixed, and incubated at 32°C in a thermal cycler for 5 min. The tube was placed in the magnet and incubated at room temperature for 5 min or until the solution became clear. The supernatant contained the equalized library at ∼100 pM, and the same amount of the 12 libraries was pooled to perform the emulsion PCR. Next, emulsion PCR was performed using the Ion OneTouch™ System and Ion OneTouch™ 200 Template Kit v2 (Thermo Fisher Scientific). Template-positive Ion Sphere™ Particles (ISPs) were enriched using Dynabeads MyOne™ Streptavidin C1 beads (Invitrogen) and were washed with Ion OneTouch Wash Solution. This process was performed on an Ion OneTouch™ ES System (Thermo Fisher Scientific). The quality of the ISPs was evaluated using a Qubit 2.0 Fluorometer (Invitrogen). The enriched ISPs were sequenced on a 314 v2 Ion Chip (12 samples per chip) using an Ion Torrent Personal Genome Machine (PGM) sequencer system (Thermo Fisher Scientific) using the Ion PGM Sequencing 200 Kit version 2 (Thermo Fisher Scientific). Sequencing was performed using 500 flow runs, which generated approximately 200 bp. The PGM sequencing run outputs were directly loaded to the Torrent Server and stored as '.dat' files. Data analysis comprising annotation of single-nucleotide variants, insertions, deletions, and splice-site alterations was performed using the Ion Reporter™ Server System (Life Technologies). Sequence data were also visually examined and verified using the Integrative Genomics Viewer (IGV). Sequencing generated an average of 302,346 reads per patient, and 96.14% of these regions were mapped to the BRCA1 and BRCA2 loci. Amplicons with coverage less than 30x on the Ion Torrent™ platform, as well as pathogenic variants, and new variants were reanalyzed by Sanger sequencing.
The complete coding regions of BRCA1 (NM_7294.3) and BRCA2 (NM_000059.3), including 50-100 base pairs (bp) of non-coding sequences flanking the 5‘ and 3‘ ends of each exon, were amplified by PCR using 33 pairs of primers (for BRCA1) (1–2) (Table 1), and 48 pairs of primers (for BRCA2) (3) (Table 2). Exons 9 (helical domain) and 20 (kinase domain) of PIK3CA (NM_006218.2) were amplified by PCR (Table 3). The PCR products were analyzed by Sanger sequencing in both the forward and reverse directions.
The reaction mixture (total volume, 20 μL) contained AmpliTaq Gold enzyme 250 U (final concentration, 0.04 U/μL) (Applied Biosystems, Foster City, CA, USA), 1× AmpliTaq Gold buffer, 1.5-3.0 mM AmpliTaq Gold magnesium chloride, 0.16 mM deoxynucleotides (Invitrogen, Carlsbad, CA, USA-AM8200), primers (0.4 μM each pair), and 50 ng DNA. PCR was performed on a Veriti® 96-well Thermal Cycler (Applied Biosystems™). The PCR cycle consisted of 1 cycle at 95°C for 10 min, 40 cycles at 94°C for 50 s, 54-66°C for 50s, 72°C for 50s, and 1 cycle at 72°C for 7 min. The BRCA2 exon 11 fragments were amplified by touchdown PCR, with annealing temperatures decreasing from 63°C to 56°C for fragments corresponding to the beginning of this exon to nucleotide 4526, and annealing temperatures from 68°C to 61°C for fragments corresponding to the end of the exon. PCR products were loaded onto a 1.5% agarose gel, stained with GelRed Nucleic Acid Stain (Biotium, Hayward, CA, USA), and evaluated. PCR products were treated with Illustra™ ExoStar™ 1-Step (GE Healthcare Bio-Sciences, Pittsburgh, PA, USA) and incubated at 37°C for 15 min, followed by incubation at 80°C for 15 min. All PCR products were sequenced in both forward and reverse directions using BigDye® Terminator v3.1 (Applied Biosystems, Foster City, CA, USA-4337456), according to the manufacturer's instructions. The final product was sequenced on a 3500 Genetic Analyzer (Applied Biosystems™) or ABI 3730 DNA Analyzer (Applied Biosystems™). Sequences obtained were analyzed using Mutation Surveyor DNA Variant Analysis Software (v3.30, SoftGenetics LLC, State College, PA, USA). All pathogenic mutations were confirmed by Sanger sequencing.
All patients were investigated for large rearrangements, specifically deletions and duplications, using the MLPA commercial kits, SALSA® MLPA® P002 BRCA1 probemix (P002-100R) and SALSA® MLPA® P045 BRCA2/CHEK2 probemix (P045-100R) (MRC-Holland, Amsterdam, The Netherlands). At first, 80 ng of genomic DNA resuspended in 2.5 μL ultrapure water was denatured for 10 min at 98°C after which 1.5 μL of the probemix mixture was added (0.75 μL of MLPA probe and 0.75 μL of MLPA buffer). The sample DNA and probemix mixture were heated at 95°C for 1 min and then incubated overnight at 60°C (17h). Afterward, ligation was performed using 1.5 μL of ligase buffer A, 1.5 μL of ligase buffer B, 0.5 μL of Ligase-65, and 12.5 μL of water and the reaction was incubated at 54°C for 15 min. The ligase was then inactivated by incubating the reaction at 98°C for 5 min. Amplification was performed by adding 5 μL of Polymerase mix (1 μL of SALSA PCR primers, 0.25 μL of SALSA polymerase, and 3.75 μL of water) and heated at 95°C for 1 min. PCR was carried out for 35 cycles (30s at 95°C, 30s at 60°C, and 60s at 72°C), followed by 20 min at 72°C on a GeneAmp 9700 Thermal Cycler (Applied Biosystems). Then, 1 μL of the PCR product was diluted 1:10 in water, mixed with 0.075 μL GeneScan™ 600 LIZ® dye Size Standard v2.0 (Applied Biosystems-4408399) and 9 μL Hi-Di Formamide (Applied Biosystems-4440753), and incubated at 80°C for 2 min on a GeneAmp 9700 Thermal Cycler (Applied Biosystems). The fragments were analyzed on an Applied Biosystems 3500 Genetic Analyzer (Applied Biosystems™) or ABI 3730 DNA Analyzer (Applied Biosystems™), and analysis was performed using Coffalyser.Net MLPA Analysis Software (MRC-Holland, Amsterdam, Netherlands). To normalize the data, at least three genomic DNA samples obtained from the peripheral blood cells of healthy donors were used as controls in each analysis. Normal values were considered when the ratio was between 0.8 and 1.2.
Variants were named according to the Human Genome Variation Society (HGVS) nomenclature (4). BRCA1 and BRCA2 variants were searched in publicly accessible databases, i.e., BRCA Share™ (5,6), BRCA Exchange (7), BRCA Mutation Database (8), and ClinVar (9); this search was performed between January and June 2020. Gene variants were evaluated using the following in silico prediction models: Polymorphism Phenotyping (PolyPhen; v2.2.2) (10), Sorting Intolerant From Tolerant (SIFT; v1.0.3) (11), Align-GVGD (12,13), Protein Variation Effect Analyzer (Provean; v1.1) (14), and Human Splicing Finder (15) to identify variants of unknown clinical significance. Minor allele frequency (MAF) was checked using the 1000 Genomes Project database (16), the Exome Aggregation Consortium (ExAC) (17,18), Global MAF dbSNP (19), Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP) (20), Genome Aggregation Database (gnomAD) (21), Trans-Omics for Precision Medicine (TOPMed) (22), and Brazilian genomic variants (ABraOM) (23).
The variants were then classified according to the recommendations of the American College of Medical Genetics and Genomics in pathogenic, likely pathogenic, benign, likely benign, and variant of uncertain significance (VUS) (24). VUS for BRCA was also checked for co-occurrence with known pathogenic mutations in the same patient. For some variants, we considered that consensus information in ≥2 databases was strong enough to classify them as benign or VUS.
- 1
N Arnold, E Gross, U Schwarz-Boeger, J Pfisterer, W Jonat, M Kiechle. A highly sensitive, fast, and economical technique for mutation analysis in hereditary breast and ovarian cancers. Hum Mutat. 1999;14(4):333–9. https://doi.org/10.1002/(SICI)1098-1004(199910)14:4<333::AID-HUMU9>3.0.CO;2-C
- 2
LS Friedman, EA Ostermeyer, CI Szabo, P Dowd, ED Lynch, SE Rowell, et al. Confirmation of BRCA1 by analysis of germline mutations linked to breast and ovarian cancer in ten families. Nat Genet. 1994;8(4):399–404. https://doi.org/10.1038/ng1294-399
- 3
TM Wagner, K Hirtenlehner, P Shen, R Moeslinger, D Muhr, E Fleischmann, et al. Global sequence diversity of BRCA2: analysis of 71 breast cancer families and 95 control individuals of worldwide populations. Hum Mol Genet. 1999;8(3):413–23. https://doi.org/10.1093/hmg/8.3.413
- 4
JT den Dunnen, R Dalgleish, DR Maglott, RK Hart, MS Greenblatt, J McGowan-Jordan, et al. HGVS Recommendations for the Description of Sequence Variants: 2016 Update. Hum Mutat. 2016;37(6):564–9. https://doi.org/10.1002/humu.22981
- 5
C Béroud, G Collod-Béroud, C Boileau, T Soussi, C Junien. UMD (Universal mutation database): a generic software to build and analyze locus-specific databases. Hum Mutat. 2000;15(1):86–94. https://doi.org/10.1002/(SICI)1098-1004(200001)15:1<86::AID-HUMU16>3.0.CO;2-4
- 6
S Caputo, L Benboudjema, O Sinilnikova, E Rouleau, C Béroud, R Lidereau, et al. Description and analysis of genetic variants in French hereditary breast and ovarian cancer families recorded in the UMD-BRCA1/BRCA2 databases. Nucleic Acids Res. 2012;40(Database issue):D992–1002. https://doi.org/10.1093/nar/gkr1160
- 7
BRCA Exchange. Available from: https://brcaexchange.org/ [Accessed June 28th, 2020]
- 8
Arup Laboratories. Available from: https://arup.utah.edu/database/BRCA/Home/BRCA1_landing.php [Accessed June 28th, 2020]
- 9
MJ Landrum, JM Lee, GR Riley, W Jang, WS Rubinstein, DM Church, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42(Database issue):D980–5. https://doi.org/10.1093/nar/gkt1113
- 10
IA Adzhubei, S Schmidt, L Peshkin, VE Ramensky, A Gerasimova, P Bork, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–9. https://doi.org/10.1038/nmeth0410-248
- 11
PC Ng, S Henikoff. Predicting deleterious amino acid substitutions. Genome Res. 2001;11(5):863–74. https://doi.org/10.1101/gr.176601
- 12
SV Tavtigian, AM Deffenbaugh, L Yin, T Judkins, T Scholl, PB Samollow, et al. Comprehensive statistical study of 452 BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. J Med Genet. 2006;43(4):295–305. https://doi.org/10.1136/jmg.2005.033878
- 13
E Mathe, M Olivier, S Kato, C Ishioka, P Hainaut, SV Tavtigian. Computational approaches for predicting the biological effect of p53 missense mutations: a comparison of three sequence analysis based methods. Nucleic Acids Res. 2006;34(5):1317–25. https://doi.org/10.1093/nar/gkj518
- 14
Y Choi, GE Sims, S Murphy, JR Miller, AP Chan. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7(10):e46688. https://doi.org/10.1371/journal.pone.0046688
- 15
FO Desmet, D Hamroun, M Lalande, G Collod-Béroud, M Claustres, C Béroud. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37(9):e67. https://doi.org/10.1093/nar/gkp215
- 16
1000 Genomes Project Consortium, GR Abecasis, A Auton, LD Brooks, MA DePristo, RM Durbin, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. https://doi.org/10.1038/nature11632
- 17
http://exac.broadinstitute.org/variant/9-139413097-T-G [Accessed June 28th, 2020]
- 18
M Lek, KJ Karczewski, EV Minikel, KE Samocha, E Banks, T Fennell, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–91. https://doi.org/10.1038/nature19057
- 19
https://pubchem.ncbi.nlm.nih.gov/
- 20
http://evs.gs.washington.edu/EVS/
- 21
Genome Aggregation Database (gnomAD). Availabel from: https://gnomad.broadinstitute.org/
- 22
Trans-Omics for Precision Medicine(TOPMed)
- 23
MS Naslavsky, GL Yamamoto, TF de Almeida, SAM Ezquina, DY Sunaga, N Pho, et al. Exomic variants of an elderly cohort of Brazilians in the ABraOM database. Hum Mutat. 2017;38(7):751–63. https://doi.org/10.1002/humu.23220
- 24
S Richards, N Aziz, S Bale, D Bick, S Das, J Gastier-Foster, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–24. https://doi.org/10.1038/gim.2015.30
No potential conflict of interest was reported.