Conflicting data from studies on the potential role of multidrug resistance 1 gene polymorphisms in inflammatory bowel disease may result from the analysis of genetically and geographically distinct populations. Here, we investigated whether multidrug resistance 1 gene polymorphisms are associated with inflammatory bowel diseases in patients from Rio de Janeiro.
METHODS:We analyzed 123 Crohn's disease patients and 83 ulcerative colitis patients to determine the presence of the multidrug resistance 1 gene polymorphisms C1236T, G2677T and C3435T. In particular, the genotype frequencies of Crohn's disease and ulcerative colitis patients were analyzed. Genotype-phenotype associations with major clinical characteristics were established, and estimated risks were calculated for the mutations.
RESULTS:No significant difference was observed in the genotype frequencies of the multidrug resistance 1 G2677T/A and C3435T polymorphisms between Crohn's disease and ulcerative colitis patients. In contrast, the C1236T polymorphism was significantly more common in Crohn's disease than in ulcerative colitis (p = 0.047). A significant association was also found between the multidrug resistance 1 C3435T polymorphism and the stricturing form of Crohn's disease (OR: 4.13; p = 0.009), whereas no association was found with penetrating behavior (OR: 0.33; p = 0.094). In Crohn's disease, a positive association was also found between the C3435T polymorphism and corticosteroid resistance/refractoriness (OR: 4.14; p = 0.010). However, no significant association was found between multidrug resistance 1 gene polymorphisms and UC subphenotypic categories.
CONCLUSION:The multidrug resistance 1 gene polymorphism C3435T is associated with the stricturing phenotype and an inappropriate response to therapy in Crohn's disease. This association with Crohn's disease may support additional pathogenic roles for the multidrug resistance 1 gene in regulating gut-microbiota interactions and in mediating fibrosis. Understanding the effects of several drugs associated with multidrug resistance 1 gene variants may aid in the selection of customized therapeutic regimens.
Inflammatory bowel diseases (IBDs) comprise Crohn's disease (CD) and ulcerative colitis (UC) and are characterized by chronic and relapsing intestinal inflammation due to an inappropriate immune response to the intestinal microbiota in a genetically predisposed individual (1). The results obtained from genome-wide association studies have contributed to the identification of distinct genetic loci implicated in IBD susceptibility, including pathways involved in pro-inflammatory cell activation (2) and autophagy (3). However, after the discovery of nucleotide-binding oligomerization domain 2/caspase recruitment domain-containing protein 15 (NOD2/CARD15) as the first susceptibility gene for CD (4,5), research on IBD has progressively shifted toward investigation of the innate immune system and the integrity of the epithelial barrier.
Among the genes regulating innate immunity is a member of the adenosine triphosphate-binding cassette superfamily, ABCB1, which is also known as multidrug resistance 1 (MDR1). MDR1 is located in an IBD susceptibility locus on chromosome 7q21 (6,7) and is also involved in epithelial integrity (8); thus, this gene has emerged as an interesting candidate for the study of IBD pathogenesis. Moreover, because the encoded product of the MDR1 gene, P-glycoprotein 170 (P-gp), is highly expressed on epithelial surfaces such as the brush borders of enterocytes (9), it has been suggested that this transmembrane efflux pump could participate in the function of the intestinal barrier, preventing the accumulation of toxins (10). In addition, proper P-gp function appears to contribute to the prevention of colon inflammation because mdr1a-knockout mice develop spontaneous colitis under specific pathogen-free conditions (11).
Several drugs routinely used in IBD therapy, including corticosteroids (12,13) and immunosuppressants, such as methotrexate (14) and cyclosporin A (15), are also substrates of P-gp. This glycoprotein functions by transporting molecules from the inner to the outer leaflet of the cell membrane. High expression of the P-gp protein was demonstrated in the peripheral blood lymphocytes and the enterocytes of patients with CD and UC who required surgical treatment after the failure of medical therapy. This result prompted the investigators to hypothesize that the lack of a response to steroids in IBD could be explained by constitutively high MDR1 expression (16). An increased efflux of steroids, mediated by P-gp, would then result in decreased concentrations of cytoplasmic steroids in enterocytes, reducing the drugs' pharmacological effectiveness (17). Nevertheless, the biological functions of these gene variants and the question of whether they can modulate the IBD phenotype are still unclear.
To date, studies on MDR1 gene polymorphisms and their potential association with IBD have provided conflicting results. Furthermore, no studies on MDR1 alleles, MDR1 genotypes and their respective frequencies have been performed in Brazilian patients with IBD. Therefore, in view of the conflicting data and the potential relevance of MDR1 gene polymorphisms to determining specific IBD behaviors, we examined the contributions of the MDR1 polymorphisms C1236T, C3435T and G2677T/A in a southeastern Brazilian population. Additionally, we investigated the relationship between genotype and clinical phenotype in IBD patients from Rio de Janeiro.
MATERIALS AND METHODSStudy populationA total of 206 patients with IBD (comprising 123 patients with CD and 83 patients with UC) were enrolled in this study from February 2009 to January 2011. The patients were regularly followed up at the Outpatient Unit for Intestinal Diseases of the Disciplina de Gastreonterologia e Endoscopia Digestiva of the Hospital Universitário Pedro Ernesto, Universidade do Estado do Rio de Janeiro (HUPE/UERJ), and of the Serviço de Gastreonterologia of the Hospital Universitário Clementino Fraga Filho, Universidade Federal do Rio de Janeiro (HUCCF/UFRJ). The diagnosis of IBD was based on established diagnostic criteria, including clinical, imaging, endoscopic and histological parameters (18). Clinicopathological data were collected from all patients, including gender, ethnicity, age, age at diagnosis, disease activity, their history of surgery related to IBD, chronic steroid use (including steroid-dependent or steroid-refractory disease) and the presence of side effects of medical treatment. For the patients with CD, the disease location was characterized as the terminal ileum (L1), colon (L2), ileocolon (L3) or upper gastrointestinal tract (L4), and the predominant disease behavior was defined as non-stricturing, non-penetrating (B1); stricturing (B2); or perforating (B3) according to the Montreal classification (19). Perianal disease was considered separately as an additional feature. CD activity assessment was based on the Harvey-Bradshaw index (20). For the patients with UC, disease extension was characterized based on the Montreal classification, using modified criteria combining ulcerative proctitis and left-sided UC (E1+E2) and considering extensive UC separately (pancolitis; E3). Disease activity was also assessed using the Clinical Colitis Activity Index (21).
DNA extraction and genotypingPeripheral blood samples were obtained from all of the participants by venipuncture and collected in EDTA tubes. Genomic DNA was isolated from peripheral blood leukocytes by proteinase-K/sodium dodecyl sulfate digestion and phenol-chloroform extraction, as described elsewhere (22). The MDR1 polymorphisms most commonly described in the literature, C1236T, G2677T and C3435T, were detected by real-time polymerase chain reaction (PCR) followed by direct sequencing. Specific primers were used for each region of interest (corresponding to exons 12, 21 and 26 of the MDR1 gene). The primers used were as follows: C1236T sense, 5′ CCTATATCCTGTGTCTGTG 3′; C1236T anti-sense, 5′ CTGTGGGGTCATAGAGCCTC 3′; G2677T sense, 5′ AGCAGGAGTTGTTGAAATGAA 3′; G2677T anti-sense, 5′ AGAGCATAGTAAGCAGTAGG 3′; C3435T sense, 5′ CGAGCACACCTGGGCATC 3′; and C3435T anti-sense, 5′ GAGGCTGCCACATGCTCCCA 3′. The genotype frequencies of the MDR1 polymorphisms were specifically analyzed in the study population of CD and UC patients.
Briefly, PCR was performed using the Rotor-Gene Q 2plex HRM (Qiagen, Limburg, Netherlands) with two channels (green and yellow). The reactions were performed in a buffer containing 0.75 mM MgCl2, 0.2 mM dNTPs and 1.0 U of Platinum Taq DNA polymerase (all from Invitrogen, Life Technologies, Carlsbad, CA, USA); 20 pmol of each primer; 200 ng of genomic DNA; and sterile, ultra-pure water, to a final volume of 50 μL. For amplification, the DNA was first denatured for 5 minutes at 94°C, and 35 cycles consisting of three steps were then performed: denaturation for 30 seconds at 92°C, annealing for 30 seconds at 60°C (exons 12 and 21) or 58°C (exon 26) and extension for 1 minute at 72°C. Subsequently, a final cycle of 10 minutes at 72°C was performed. The PCR products were then purified with the Illustra GFX™ PCR DNA and Gel Band Purification Kit according to the manufacturer's protocol (GE Healthcare, Buckinghamshire, UK).
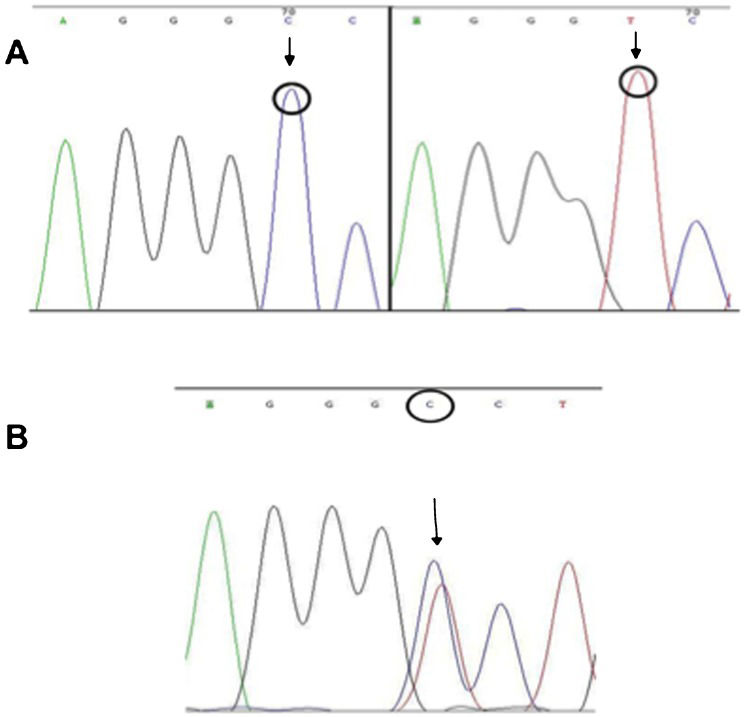
The sequencing reactions were performed using the ET Dye Terminator Cycle Sequencing Kit (GE Healthcare) according to the manufacturer's protocol. The primers used were the same as those employed in the PCR (Table1). For each product, eight sequencing reactions were performed: four with sense oligonucleotides and four with anti-sense oligonucleotides. The sequencing reactions were then analyzed using the MegaBACE 1000 automatic sequencer (GE Healthcare), and the sequences were analyzed using Chromas software (http://www.technelysium.com.au/chromas.html, accessed on March 19th, 2011) (Figure1).
MDR1 C1236T, G2677T/A and C3435T genotypes and allele frequencies.
| Polymorphism | CHz | HTz | RHz | n | Allele frequency | χ2 | p-value | |
|---|---|---|---|---|---|---|---|---|
| C | T | |||||||
| C1236T | C:C | C:T | T:T | |||||
| Observed | 90 | 89 | 27 | 206 | 0.65 | 0.35 | 0.45 | 0.50 |
| Expected | 88 | 93 | 25 | |||||
| G2677T/A | G:G | G:T/A | T/A:T/A | |||||
| Observed | 81 | 105 | 20 | 206 | 0.65 | 0.35 | 2.83 | 0.09 |
| Expected | 87 | 94 | 25 | |||||
| C3435T | C:C | C:T | T:T | |||||
| Observed | 92 | 83 | 31 | 206 | 0.65 | 0.35 | 2.80 | 0.09 |
| Expected | 87 | 94 | 25 |
CHz, common homozygote; HTz, heterozygote; RHz, rare homozygote.
Electropherogram 1236. The wild-type sequence agggCc (left) and the polymorphism in which cytosine is exchanged for thymine (agggTc) (right) (A). Electropherogram showing a polymorphism (C1236T) with overlapping cytosine and thymine curves, indicating a heterozygous individual (B).
Tests for Hardy-Weinberg equilibrium were performed using Genepop software (Genepop web version 3.1). The 5% significance level for one degree of freedom is 3.84, and because the qui-square value was less than this, the null hypothesis that the population was in Hardy-Weinberg equilibrium was not rejected.
For all other data evaluation, we used SPSS 17.0 software (SPSS Inc., Chicago, IL, USA). The distribution of individual characteristics was evaluated by simple descriptive statistics. Differences among the distributions of selected variables were evaluated using the chi-square test for categorical data. All tests were two-tailed, and p-values of less than 0.05 were considered statistically significant. Genotype-phenotype associations were assessed using odds ratios (ORs) calculated for the minor allele at each single-nucleotide polymorphism (SNP). Multiple logistic regression models were also used to explore the effect of genotype on the phenotypic variables, with phenotype status as the dependent variable.
Ethical considerationsThis study was approved by the Ethical Committees of the University Hospital Pedro Ernesto, Universidade do Estado do Rio de Janeiro, and of the University Hospital Clementino Fraga Filho, Universidade Federal do Rio de Janeiro. Informed consent was obtained from all subjects. The study protocol was in accordance with the ethical principles for medical research involving human subjects described in the Declaration of Helsinki.
RESULTSThe CD group consisted of 49 men and 74 women with a mean age of 39.8 years (range: 11–80 years). In total, 51 were classified as white, whereas 72 were classified as non-white. The mean duration of CD was 8.8 years (range: 0.5–41 years). The UC group consisted of 40 men and 43 women with a mean age of 45.6 years (range: 21–73 years). Of these individuals, 31 were classified as white, and 52 were classified as as non-white. The mean duration of UC was 7.5 years (range: 0.2–27 years).
The distributions of the selected MDR1 gene polymorphisms (C1236T, G2677T/A and C3435T) are shown in Table1 and demonstrate that the respective allele frequencies were in Hardy-Weinberg equilibrium in the study population.
Subsequently, we investigated the distribution of each polymorphic genotype in the CD and UC patient groups (Table2). The MDR1 G2677T/A and C3435T genotypes were similar between CD and UC patients (p = 0.477 and p = 0.712, respectively). However, the homozygous C1236T genotype was significantly more prevalent among CD patients compared with UC patients (p = 0.047).
Analysis of MDR1 gene polymorphisms for the differential diagnosis of inflammatory bowel disease.
| MDR1 SNP | CHz | HTz | RHz | p-value |
|---|---|---|---|---|
| C1236T | C:C | C:T | T:T | |
| CD n = 123) | 50 | 51 | 22 | 0.047 |
| UC (n = 83) | 40 | 38 | 5 | |
| G2677T/A | G:G | G:T/A | T/A:T/A | |
| CD n = 123) | 50 | 59 | 14 | 0.477 |
| UC (n = 83) | 31 | 46 | 6 | |
| C3435T | C:C | C:T | T:T | |
| CD n = 123) | 52 | 52 | 19 | 0.712 |
| UC (n = 83) | 40 | 31 | 12 |
MDR1, multidrug resistance protein 1; SNP, single-nucleotide polymorphism; CD, Crohn's disease; UC, ulcerative colitis; CHz, common homozygous; HTz, heterozygous; RHz, rare homozygous. The data were analyzed using the Pearson chi-square test.
The association of the different polymorphic genotypes with the phenotypic characteristics of CD and UC was also investigated. Tables3, 4 and 5 show the genotype frequencies of the SNPs in different subgroups of patients with CD. A significant positive association was found between the MDR1 homozygous C3435T polymorphism and the stricturing form of CD (p = 0.009). Interestingly, a tendency toward a negative association with the penetrating form of the disease was identified for the same SNP (p = 0.094). Among the 35 patients who had stricturing disease, 11 had the homozygous C3435T polymorphism, whereas among the 44 patients with penetrating disease, only three presented this genotype. Furthermore, the homozygous C3435T polymorphism was associated with chronic steroid use (steroid-dependent/refractory) (p = 0.010) (Table5). A significant association with disease behavior was also found between the MDR1 heterozygous G2677T/A polymorphism and the non-stricturing, non-penetrating form of CD (p = 0.033; Table4). In contrast, no significant associations were found between the MDR1 C1236T polymorphism and specific CD subphenotypes (Table3). Furthermore, no significant association was found between the MDR1 gene polymorphisms and UC subphenotypic categories (Supplementary Tables S1, S2 and S3).
Genotype-phenotype associations of the MDR1 SNP C1236T in patients with Crohn's disease.
| MDR1 SNP C1236T | CHz | HTz | OR | p-value | RHz | OR | p-value |
|---|---|---|---|---|---|---|---|
| Gender | |||||||
| Male (n = 49, 39.8%) | 20 | 19 | 0.89 | 0.777 | 10 | 1.25 | 0.665 |
| Female (n = 74, 60.2%) | 30 | 32 | 12 | ||||
| Ethnicity | |||||||
| White (n = 51, 41.5%) | 20 | 22 | 1.14 | 0.749 | 9 | 1.04 | 0.942 |
| Non-white (n = 72, 58.5%) | 30 | 29 | 13 | ||||
| Age at diagnosis | |||||||
| <40 (A1) (n = 62, 50.4%) | 24 | 31 | 1.68 | 0.197 | 7 | 0.51 | 0.201 |
| ≥40 (A2) (n = 61, 49.6%) | 26 | 20 | 15 | ||||
| Disease location | |||||||
| Terminal ileum (L1) (n = 22, 17.9%) | 10 | 11 | 1.10 | 0.846 | 1 | 0.19 | 0.093 |
| Colon (L2) (n = 58, 47.1%) | 23 | 21 | 0.82 | 0.625 | 14 | 2.05 | 0.168 |
| Ileocolon (L3) (n = 38, 30.9%) | 14 | 18 | 1.40 | 0.431 | 6 | 0.96 | 0.949 |
| Upper GI (L4) (n = 5, 4.1%) | 3 | 1 | 0.31 | 0.298 | 1 | 0.75 | 0.804 |
| Disease behavior | |||||||
| NS/NP (B1) (n = 44, 35.8%) | 17 | 19 | 1.15 | 0.732 | 8 | 1.11 | 0.846 |
| Stricturing (B2) (n = 35, 28.4%) | 15 | 14 | 0.88 | 0.777 | 6 | 0.88 | 0.814 |
| Penetrating (B3) (n = 44, 35.8%) | 18 | 18 | 0.97 | 0.941 | 8 | 1.02 | 0.976 |
| Disease activity | |||||||
| Moderate/severe (n = 24, 19.5%) | 8 | 12 | 1.62 | 0.342 | 4 | 1.17 | 0.819 |
| Mild/remission (n = 99, 80.5%) | 42 | 39 | 18 | ||||
| Surgery due to CD | |||||||
| Yes (n = 45, 36.6%) | 23 | 14 | 0.44 | 0.053 | 8 | 0.67 | 0.447 |
| No (n = 78, 63.4%) | 27 | 37 | 14 | ||||
| Perianal disease | |||||||
| Yes (n = 37, 30.1%) | 17 | 15 | 0.81 | 0.620 | 5 | 0.57 | 0.338 |
| No (n = 86, 69.9%) | 33 | 36 | 17 | ||||
| Chronic steroid use | |||||||
| Yes (n = 33, 26.8%) | 10 | 15 | 1.67 | 0.273 | 8 | 2.29 | 0.139 |
| No (n = 90, 73.2%) | 40 | 36 | 14 | ||||
| Side effects of medication | |||||||
| Yes (n = 39, 31.7%) | 15 | 18 | 1.27 | 0.570 | 6 | 0.88 | 0.814 |
| No (n = 84, 68.3%) | 35 | 33 | 16 |
CHz, common homozygote (C:C); HTz, heterozygote (C:T); RHz, rare homozygote (T:T); NS/NP, non-stricturing, non-penetrating; OR, odds ratio. All comparisons were performed in relation to the CHz group.
Genotype-phenotype associations of the MDR1 SNP C3435T in patients with Crohn's disease.
| MDR1 SNP C3435T | CHz | HTz | OR | p-value | RHz | OR | p-value |
|---|---|---|---|---|---|---|---|
| Gender | |||||||
| Male (n = 49, 39.8%) | 22 | 18 | 0.72 | 0.420 | 9 | 1.23 | 0.703 |
| Female (n = 74, 60.2%) | 30 | 34 | 10 | ||||
| Ethnicity | |||||||
| White (n = 51, 41.5%) | 22 | 21 | 0.92 | 0.842 | 8 | 0.99 | 0.988 |
| Non-white (n = 72, 58.5%) | 30 | 31 | 11 | ||||
| Age at diagnosis | |||||||
| <40 (A1) (n = 62, 50.4%) | 25 | 27 | 1.17 | 0.695 | 10 | 1.20 | 0.734 |
| ≥40 (A2) (n = 61, 49.6%) | 27 | 25 | 9 | ||||
| Disease location | |||||||
| Terminal ileum (L1) (n = 22, 17.9%) | 8 | 10 | 1.31 | 0.604 | 4 | 1.47 | 0.572 |
| Colon (L2) (n = 58, 47.1%) | 25 | 25 | 1.00 | 1.000 | 8 | 0.79 | 0.655 |
| Ileocolon (L3) (n = 38, 30.9%) | 16 | 16 | 1.00 | 1.000 | 6 | 1.04 | 0.948 |
| Upper GI (L4) (n = 5, 4.1%) | 3 | 1 | 0.32 | 0.307 | 1 | 0.91 | 0.934 |
| Disease behavior | |||||||
| NS/NP (B1) (n = 44, 35.8%) | 20 | 19 | 0.92 | 0.839 | 5 | 0.57 | 0.343 |
| Stricturing (B2) (n = 35, 28.4%) | 13 | 11 | 0.80 | 0.641 | 11 | 4.13 | 0.009* |
| Penetrating (B3) (n = 44, 35.8%) | 19 | 22 | 1.27 | 0.547 | 3 | 0.33 | 0.094 |
| Disease activity | |||||||
| Moderate/severe (n = 24, 19.5%) | 7 | 11 | 1.72 | 0.299 | 6 | 2.97 | 0.080 |
| Mild/remission (n = 99, 80.5%) | 45 | 41 | 13 | ||||
| Surgery due to CD | |||||||
| Yes (n = 45, 36.6%) | 20 | 20 | 1.00 | 1.000 | 5 | 0.57 | 0.342 |
| No (n = 78, 63.4%) | 32 | 32 | 14 | ||||
| Perianal disease | |||||||
| Yes (n = 37, 30.1%) | 17 | 16 | 0.92 | 0.833 | 4 | 0.55 | 0.341 |
| No (n = 86, 69.9%) | 35 | 36 | 15 | ||||
| Chronic steroid use | |||||||
| Yes (n = 33, 26.8%) | 11 | 12 | 1.12 | 0.813 | 10 | 4.14 | 0.010* |
| No (n = 90, 73.2%) | 41 | 40 | 9 | ||||
| Side effects of medication | |||||||
| Yes (n = 39, 31.7%) | 13 | 22 | 2.20 | 0.062 | 4 | 0.80 | 0.730 |
| No (n = 84, 68.3%) | 39 | 30 | 15 |
CHz, common homozygous (C:C); HTz, heterozygous (C:T); RHz, rare homozygous (T:T); NS/NP, non-stricturing, non-penetrating; OR, odds ratio. All comparisons were performed in relation to the CHz group.
Genotype-phenotype associations of the MDR1 SNP G2677T/A in patients with Crohn's disease.
| MDR1 SNP G2677T/A | CHz | HTz | OR | p-value | RHz | OR | p-value |
|---|---|---|---|---|---|---|---|
| Gender | |||||||
| Male (n = 49, 39.8%) | 17 | 26 | 1.53 | 0.284 | 6 | 1.46 | 0.541 |
| Female (n = 74, 60.2%) | 33 | 33 | 8 | ||||
| Ethnicity | |||||||
| White (n = 51, 41.5%) | 19 | 26 | 1.29 | 0.521 | 6 | 1.22 | 0.742 |
| Non-white (n = 72, 58.5%) | 31 | 33 | 8 | ||||
| Age at diagnosis | |||||||
| <40 (A1) (n = 62, 50.4%) | 24 | 31 | 1.20 | 0.636 | 7 | 1.08 | 0.894 |
| ≥40 (A2) (n = 61, 49.6%) | 26 | 28 | 7 | ||||
| Disease location | |||||||
| Terminal ileum (L1) (n = 22, 17.9%) | 10 | 10 | 0.82 | 0.682 | 2 | 0.67 | 0.628 |
| Colon (L2) (n = 58, 47.1%) | 27 | 26 | 0.67 | 0.301 | 5 | 0.47 | 0.226 |
| Ileocolon (L3) (n = 38, 30.9%) | 11 | 21 | 1.96 | 0.120 | 6 | 2.66 | 0.118 |
| Upper GI (L4) (n = 5, 4.1%) | 2 | 2 | 0.84 | 0.865 | 1 | 1.85 | 0.623 |
| Disease behavior | |||||||
| NS/NP (B1) (n = 44, 35.8%) | 13 | 27 | 2.40 | 0.033* | 4 | 1.14 | 0.847 |
| Stricturing (B2) (n = 35, 28.4%) | 16 | 13 | 0.60 | 0.240 | 6 | 1.59 | 0.449 |
| Penetrating (B3) (n = 44, 35.8%) | 21 | 19 | 0.66 | 0.290 | 4 | 0.55 | 0.362 |
| Disease activity | |||||||
| Moderate/severe (n = 24, 19.5%) | 8 | 12 | 1.34 | 0.559 | 4 | 2.10 | 0.286 |
| Mild/remission (n = 99, 80.5%) | 42 | 47 | 10 | ||||
| Surgery due to CD | |||||||
| Yes (n = 45, 36.6%) | 23 | 17 | 0.48 | 0.063 | 5 | 0.65 | 0.493 |
| No (n = 78, 63.4%) | 27 | 42 | 9 | ||||
| Perianal disease | |||||||
| Yes (n = 37, 30.1%) | 15 | 17 | 0.94 | 0.892 | 5 | 1.30 | 0.683 |
| No (n = 86, 69.9%) | 35 | 42 | 9 | ||||
| Chronic steroid use | |||||||
| Yes (n = 33, 26.8%) | 12 | 14 | 0.99 | 0.973 | 7 | 3.17 | 0.060 |
| No (n = 90, 73.2%) | 38 | 45 | 7 | ||||
| Side effects of medication | |||||||
| Yes (n = 39, 31.7%) | 17 | 20 | 1.00 | 0.991 | 2 | 0.32 | 0.153 |
| No (n = 84, 68.3%) | 33 | 39 | 12 |
CHz, common homozygous (G:G); HTz, heterozygous (G:T/A); RHz, rare homozygous (T/A:T/A); NS/NP, non-stricturing, non-penetrating; OR, odds ratio. All comparisons were performed in relation to the CHz group.
In this study, we present information on the genotype frequencies of the MDR1 C1236T, G2677T/A and C3435T polymorphisms in patients with CD or UC as well as potential determination of the phenotypic features of IBD, in a southeastern Brazilian population from Rio de Janeiro. In particular, we found that the C1236T polymorphism was significantly more common in CD patients than in UC patients. Regarding CD phenotypes, a significant association was detected between the MDR1 C3435T polymorphism and the stricturing form of the disease. In addition, a positive association between the C3435T polymorphism and the chronic use of steroids was identified in CD patients. We also found positive and negative trends regarding specific phenotypes and both heterozygous and homozygous polymorphisms, suggesting that MDR1 gene variants may determine both susceptibility to and protection against CD.
Various MDR1 gene polymorphisms have been reported thus far, and the C3435T polymorphism has been the most well studied in IBD. Similar to the results of the current study, associations between MDR1 gene polymorphisms have been reported in refractory CD and, to a lesser extent, in UC in a Slovenian population (23). In another study performed in a large cohort in the United States, investigators observed a significant association between CD and a missense polymorphism in exon 21 (G2677T/C; Ala893Ser/Thr), which was thought to be related to altered transporter and/or gene expression activity (24). In a case-control study in Spain, a significant association between the MDR1 C3435T polymorphism and CD was characterized, in addition to the identification of the CD susceptibility haplotype 2677T/C3435 (25). Moreover, in an Italian study, investigators found a significant association between the C3435T SNP and the ileocolonic localization of CD, whereas the same polymorphism appeared to be negatively associated with a positive family history and arthritis in CD patients (26). In a large case-control cohort study in the United Kingdom, investigators associated the MDR1 SNPs C3435T and G2677T/A with an increased risk of developing UC, including an influence on disease behavior (27). In addition, in contrast to the results of the current study, case-control studies in European cohorts of patients from Germany (28) and Scotland (29) also suggested a potential association between the MDR1 SNP C3435T and UC. Nevertheless, in another Italian study, the investigated polymorphisms in the MDR1 gene had no significant role in disease susceptibility or the response to medical therapy in IBD (30).
The discrepancies among these studies may be attributed to not only different study designs, sample sizes and selection of controls but also the distinct patient populations. In fact, meta-analyses have been performed to attempt to overcome the heterogeneity of studies involving MDR1 SNPs. Of note, certain studies have revealed that the allele frequencies of the three main variants differ considerably among distinct populations. A significant association of the 3435T allele with UC was confirmed in a meta-analysis (27), and in another study, the 3435T allele and the 3435TT genotype were demonstrated to be significantly associated with UC, but not with CD (31). Differences in the C3435T SNP allele frequency have also been detected, with an increased frequency of the C allele (wild type) in African populations compared with Caucasian and Asian populations (32). In an Asiatic study, investigators found distinct haplotype profiles and linkage disequilibrium at the MDR1 gene locus in all three ethnic groups enrolled in the study (33). In a recent study that was also performed in Rio de Janeiro, Brazil, matching the recruitment area and ethnicity of our IBD patients, 278 healthy individuals were analyzed regarding the genotype and allele frequencies of MDR1 gene polymorphisms. The investigators found a peculiar variant distribution, with significant differences between C1236C and C3435T and also between C1236C and C3435C, which differed from the results obtained for several other ethnic groups (34). In this context, it must be emphasized that data from potential source populations, such as Europeans or Africans, cannot be deemed representative of the Brazilian genotype and allele frequencies due to the marked heterogeneity and the extensive admixture of the Brazilian population (35,36). Taken together with the results of our study, these observations highlight the critical importance of analyzing MDR1 gene polymorphisms in Brazilians, and particularly in Brazilian IBD patients.
Potential genotype-phenotype associations related to the MDR1 gene have been investigated, with contradictory results. In the present study, we report positive associations between the C3435T polymorphism and both the chronic use of steroids and disease activity in CD patients. Similar to our results, an association between MDR1 variants and corticosteroid dependence was also reported in children with CD in Canadian tertiary centers (37). In a British study on IBD, the 2677T allele was increased in UC cases, and the TT genotype was strongly associated with disease severity and the use of steroids in UC (27). In contrast, in a large cohort of IBD patients using steroids, both the C3435T and the G2677T/A polymorphisms were evaluated, but no significant differences were found within subgroups or among subgroups. Additionally, MDR1 genotypes were not found to influence the response to therapy (30). In another study, the expression and function of MDR1 in intraepithelial, lamina propria and peripheral blood lymphocytes were decreased in UC patients compared with CD patients and healthy controls (38). In accordance with this finding, the tissue expression of a number of detoxification genes and ABC transporters, including MDR1, was shown to be markedly downregulated in UC patients, supporting the notion that a defective mucosal detoxification system could predispose a patient to intestinal inflammation (39). Indeed, the effects of corticosteroids and other medications used in IBD on MDR1 expression have not been fully established, and the question of whether the difference in P-gp expression reflects a primary defect or occurs secondary to therapy, influencing the response to treatment, remains to be clarified.
The stricturing form of CD, also known as the fibrostenotic form of CD, has been previously associated with NOD2 variants and small bowel involvement in patients with CD (40). However, another study demonstrated that the fibrostenotic phenotype of CD was significantly associated with NOD2 gene variants and also with a high titer of antibodies against oligomannan, OmpC, I2 and Cbir, regardless of disease location (41). These results support the notion that altered innate immunity may synergize with a loss of tolerance to microbial antigens and with the adaptive immune response, thus favoring a specific CD phenotype. In contrast to studies reported thus far on MDR1 gene polymorphisms in IBD, another novel genotype-phenotype association found in our study was the significant positive association between the C3435T SNP and the stricturing form of CD. Although the mechanism by which an MDR1 SNP can determine a specific phenotype has yet to be determined, certain evidence indicates that MDR1 participates in a complex biological network with multiple physiologically relevant mediators and pathways, including pro-inflammatory cytokines (42), endotoxin-induced inflammation (43), transcription factors such as NF-kB (44,45) and cyclooxygenases (46), which can modulate MDR1 expression and activity at different levels. Hence, these intricate mechanisms reinforce a key role for P-gp in drug bioavailability and epithelial homeostasis in the context of inflammation and infection. However, the contribution of the MDR1 gene to the response to medications and possibly to IBD susceptibility or phenotypes, including the stricturing form of CD, needs further clarification.
In conclusion, the results of this study indicate that the rare homozygous MDR1 gene polymorphism C3435T is associated not only with the stricturing phenotype but also with an inappropriate response to therapy in a population of CD patients from Rio de Janeiro. The relationship with the CD phenotype supports the existence of additional roles for MDR1 in specific mechanisms underlying CD pathogenesis, such as the control of gut-microbiota interactions and the regulation of fibrosis. Furthermore, understanding the effects of several drugs associated with these MDR1 variants may aid in the selection of customized therapeutic regimens.
ACKNOWLEDGMENTSThe authors wish to thank the Brazilian research foundations CNPq and FAPERJ for their financial support. All authors approved the final version of this article.
AUTHOR CONTRIBUTIONSCarvalho AT, Froes R, Esberard B and Carneiro AJ participated in study design, patient selection and follow-up, data collection and interpretation and manuscript preparation. Grinman AB, Neto PN, Santos J, Raposo D and Simao T performed the laboratory experiments, technical troubleshooting and data collection and interpretation. Luiz RR performed statistical analysis and data interpretation. Pinto LF and Souza HS interpreted the data, obtained funding, performed statistical analysis, wrote the manuscript and critically reviewed the manuscript.
No potential conflict of interest was reported.