The differential diagnosis of B-cell lymphoproliferative processes remains a challenge for pathologists, dermatologists and oncologists, despite advances in histology, immunohistochemistry and molecular biology.
OBJECTIVE:Evaluate aid and limitations of clonality analysis in the diagnosis of primary cutaneous B-cell lymphomas and B-cell pseudolymphomas.
METHODS:This study included 29 cases of B-cell lymphoproliferative processes classified as primary cutaneous B-cell lymphomas (13), B-cell pseudolymphomas (6) and inconclusive cases (10) using histology and immunohistochemistry. The clonality analysis was performed by polymerase chain reaction analysis of immunoglobulin light chain and heavy chain rearrangements.
RESULTS:DNA quality was shown to be generally poor; eight samples were inadequate for polymerase chain reaction analysis. The results showed monoclonality in eight of the primary cutaneous B-cell lymphomas and polyclonality in four of the B-cell pseudolymphomas. In addition, monoclonality was shown in two of the inconclusive cases by histology and immunohistochemistry, demonstrating the utility of polymerase chain reaction as an ancillary diagnostic tool for primary cutaneous B-cell lymphomas.
DISCUSSION:The low quality DNA extracted from these cases demanded the use of an IgH protocol that yielded small fragments and IgK. Both methods used together improved detection.
CONCLUSION:Use of the two protocols, immunoglobulin heavy chain FR3-trad and immunoglobulin light chain-Kappa Biomed protocols for clonality analysis improved diagnostic accuracy.
B-cell lymphoproliferative processes, including primary cutaneous B-cell lymphomas (PCBCLs) and B-cell pseudolymphomas (B-PSLs), have been exhaustively studied in the last few decades.1,2 Nevertheless, differential diagnosis between the two entities remains a challenge for dermatopathologists, hematologists, dermatologists and oncologists, despite advances in histology, immunohistochemistry and molecular biology.3–6
The clinical history and physical examination contribute to the differential diagnosis of PCBCL; however, the diagnostic investigation is largely composed of histological and immunohistochemical studies.7,8 Molecular biology techniques have been described as important ancillary diagnostic tools in difficult cases.9,10 Recently, analysis of clonality through investigation of immunoglobulin light and heavy chain gene rearrangements has been used to supplement the diagnosis of PCBCL.9,11,12 Usually, polymerase chain reaction (PCR) is the molecular technique of choice due to higher sensitivity and lower cost when compared with Southern blot. As an additional advantage, this tool offers the possibility to work with formalin-fixed and paraffin-embedded tissue.12–14
PCR has been very useful in the diagnosis of PCBCL when used to investigate rearrangement of the IgH gene located on chromosome 14 (14q32).15,16 The rearrangement of IgH genes is the first rearrangement event to take place in the differentiation of B cells, preceding that of the IgL genes, and it is present in the majority of PCBCLs.17 As several regions (FR3, FR2, and FR1) of IgH genes can be evaluated, a number of protocols have been developed. These include the Biomed-2 protocols and more traditional ones.14,18,19
Sensitivity can vary from less than 20% to more than 90%, depending on the employed method. 9,13,20 In some cases, it is not possible to amplify the IgH gene because primers fail to bind to the rearrangement that occurred in the neoplastic B lymphocytes or an inadequate DNA sample. Rearrangement of the IgL-K (2q12) and IgL lambda (22q11) genes can also be investigated and may increase the diagnostic sensitivity up to 93%, particularly when combined with IgH analysis.11, 20, 21
In the present study, the impact of the analysis of IgH and IgL-K gene rearrangement in the diagnosis of B-cell lymphoproliferative cutaneous processes and the limitations of these methods are assessed.
DIAGNOSTIC METHODTwenty-nine patients with B-cell lymphoproliferative processes were evaluated. Clinical data were retrieved from the clinical records, and all cases were classified according to the histological and immunohistochemical characteristics. We included cases diagnosed by two dermatopathologists who were experts in cutaneous lymphoma without prior knowledge of the clinical setting. The pathologists established a diagnosis of PCBCL (13 cases) or B-PSL (6 cases). Whenever there was a discrepancy between the diagnoses of both pathologists or when they could not define the diagnosis, the case was considered inconclusive (10 cases) (Table 1). When the diagnosis of PCBCL was established, the cases were classified according to the WHO-EORTC 2005 Classification.22
Clinical, histological and immunohistochemical data for 29 cases with B-cell lymphoproliferative processes
| I | G | A | Dist | Localization | Morphology | Diam | N | HE | IHC | IHC | HE+IHC | L, M, V | Stage | Status |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | M | 54 | reg | face, neck, trunk, back | papule, nodule | 1 | 10 | PCBCL | Anti-CD20 + CD10+, BCL-2−, CD23− | PCBCL | PCBCL | Y,Y,Y | system | DWD (36) |
| 2 | F | 57 | loc | back | plaque | 6 | 2 | INC | Anti-CD20 + CD10 −, BCL-2 +, CD23+ | INC | INC | N,N,N | cut ativ | AWD (78) |
| 3 | M | 76 | loc | LL | nodule, plaque | 16 | 1 | PCBCL(+) | Anti-CD20 + CD10 −, BCL-2 +, CD23− | B-PSL(−) | PCBCL | N,N,N | cut ativ | DWD (20) |
| 4 | F | 63 | diss | UL, back, palate | nodule, plaque | 10 | 10 | PCBCL(+) | Anti-CD20 + CD10 +, BCL-2 +, CD23+ | B-PSL(+) | INC | N,N,N | cut ativ | AWD (182) |
| 5 | M | 50 | loc | anterior trunk | plaque | 5 | 1 | INC(−) | Anti-CD20 + CD10 +, BCL-2 −, CD23+ | B-PSL(+) | B-PSL | N,N,N | cut ativ | AWD (135) |
| 6 | M | 71 | reg | face, neck, trunk, back | nodule, plaque | 3 | 20 | B-PSL(−) | Anti-CD20 + CD10 −, BCL-2 +, CD23+ | INC(+) | INC | N,N,N | cut ativ | DWD (32) |
| 7 | M | 67 | loc | Back | plaque | 3 | 1 | B-PSL(−) | Anti-CD20 + CD10 +, BCL-2 +, CD23− | PCBCL(+) | PCBCL | N,N,N | cut ativ | AWD (66) |
| 8 | F | 36 | reg | UL e LL | papule, nodule | 2 | 15 | B-PSL | Anti-CD20 + CD10 −, BCL-2 +, CD23− | B-PSL | B-PSL | N,N,N | cut ativ | AWD (43) |
| 9 | F | 37 | diss | anterior trunk, back, LL | papule, nodule, plaque | 2 | 20 | PCBCL | Anti-CD20 + CD10 −, BCL-2 −, CD23− | PCBCL | PCBCL | N,N,N | cut ativ | AWD (172) |
| 10 | F | 62 | loc | back | nodule | 1 | 2 | PCBCL(+) | Anti-CD20 + CD10 −, BCL-2 +, CD23+ | B-PSL(+) | INC | N,N,N | cut rem | AND (199) |
| 11 | F | 43 | loc | face | papule, nodule | 0,5 | 5 | B-PSL(−) | Anti-CD20 + CD10 +, BCL-2 −, CD23+ | PCBCL(+) | PCBCL | N,N,N | cut rem | AND (138) |
| 12 | F | 26 | loc | back | nodule | 8 | 1 | INC | Anti-CD20 + CD10 −, BCL-2 +, CD23+ | INC | INC | N,N,N | cut rem | AND (33) |
| 13 | M | 74 | loc | anterior trunk | papule, plaque | 5 | 7 | PCBCL | Anti-CD20 + CD10 +, BCL-2 −, CD23+ | PCBCL | PCBCL | N,N,N | cut ativ | AWD (42) |
| 14 | M | 29 | loc | back | papule, nodule | 2 | 8 | PCBCL | Anti-CD20 + CD10 −, BCL-2 −, CD23− | PCBCL | PCBCL | N,N,N | cut rem | AND (179) |
| 15 | M | 40 | loc | back | plaque | 15 | 1 | PCBCL | Anti-CD20 + CD10 −, BCL-2 +, CD23− | PCBCL | PCBCL | N,N,N | cut rem | AND (101) |
| 16 | F | 40 | loc | UL | papule | 1 | 3 | INC(−) | Anti-CD20 + CD10 −, BCL-2 +, CD23+ | B-PSL(+) | B-PSL | N,N,N | cut rem | AND (47) |
| 17 | F | 62 | loc | gluteal region | nodule | 3 | 1 | INC | Anti-CD20 + CD10 −, BCL-2 +, CD23+ | INC | INC | N,N,N | cut ativ | AWD (102) |
| 18 | F | 32 | loc | UL | nodule | 1 | 1 | B-PSL | Anti-CD20 + CD10 −, BCL-2 +, CD23+ | B-PSL | B-PSL | N,N,N | cut rem | AND (102) |
| 19 | M | 65 | loc | scalp | plaque | 5 | 1 | PCBCL | Anti-CD20 + CD10 +, BCL-2 −, CD23+ | PCBCL | PCBCL | N,N,N | cut ativ | AWD (36) |
| 20 | M | 85 | loc | face | papule, nodule | 5 | 3 | B-PSL | Anti-CD20 + CD10 −, BCL-2 −, CD23− | B-PSL | B-PSL | N,N,N | cut ativ | AWD (66) |
| 21 | M | 51 | loc | face | papule, nodule | 3 | 3 | B-PSL(−) | Anti-CD20 + CD10 +, BCL-2 +, CD23+ | INC(+) | INC | N,N,N | cut ativ | AWD (96) |
| 22 | F | 68 | loc | scalp | nodule | 3 | 2 | PCBCL | Anti-CD20 + CD10 −, BCL-2 −, CD23− | PCBCL | PCBCL | N,N,N | cut ativ | AWD (62) |
| 23 | M | 64 | loc | back | nodule | 5 | 1 | PCBCL(+) | Anti-CD20 + CD10 −, BCL-2 +, CD23+ | B-PSL(+) | INC | N,N,N | cut rem | AND (60) |
| 24 | M | 23 | loc | UL | plaque | 1 | 1 | PCBCL(+) | Anti-CD20 + CD10 +, BCL-2 +, CD23+ | B-PSL(+) | INC | N,N,N | cut ativ | AWD (96) |
| 25 | M | 45 | diss | trunk, UL, LL | papule, nodule | 2 | 9 | PCBCL(+) | Anti-CD20 + CD10 −, BCL-2 +, CD23− | INC(−) | PCBCL | N,N,N | cut ativ | AWD (40) |
| 26 | M | 36 | diss | trunk, UL, LL | macula, papule, plaque | 3 | 6 | INC | Anti-CD20 + CD10 +, BCL-2 +, CD23+ | INC | INC | N,N,N | cut ativ | AWD (92) |
| 27 | M | 45 | loc | scalp | nodule | 1 | 1 | PCBCL | Anti-CD20 + CD10 −, BCL-2 +, CD23− | PCBCL | PCBCL | N,N,N | cut ativ | AWD (50) |
| 28 | M | 30 | loc | scalp | nodule | 2 | 1 | PCBCL | Anti-CD20 + CD10 −, BCL-2 +, CD23− | PCBCL | PCBCL | N,N,N | cut rem | AND (60) |
| 29 | M | 31 | loc | scalp | nodule | 2 | 1 | B-PSL | Anti-CD20 + CD10 −, BCL-2 +, CD23+ | B-PSL | B-PSL | N,N,N | cut rem | AND (55) |
I: identification number of the case; G: gender; A: age in years; Dist: distribution of the cutaneous lesions; reg: regional; loc: located; diss: disseminated; UL: upper limbs; LL: lower limbs; Diam: diameter of the lesions in cm; N: number of lesions; HE: hematoxylin-eosin; IHC: immunohistochemistry results; (+): positive or important role in the diagnostic decision; (−) negative or less important role in the diagnostic decision; HE+IHC: diagnosis based on HE and IHC; L: lymph node involvement; M: bone marrow involvement; V: visceral involvement; Y: yes; N: no; system: systemic; cut ativ: cutaneous in activity; cut rem: cutaneous in remission; Status (period of follow up in months); DWD: dead with active disease; AWD: alive with active disease; AND: alive with (no) active disease.
Clonality was investigated by IgH gene rearrangement using protocols for IgH FR2 (Biomed IgH tube B), IgH FR3 (Biomed IgH tube C) and a traditional method for IgH FR3 (IgH-trad), which yields smaller fragments than the Biomed protocols.14,23 The investigation of immunoglobulin light chain (IgL) gene rearrangement was carried out using the IgL Kappa protocols (Biomed Igkappa tubes A and B).
DNA extraction (crude lysate method): Five sections of 10 μm formalin-fixed and paraffin-embedded cutaneous biopsies were deparaffinized in xylene and ethanol. Tissue digestion was carried out in 100 μl of a solution containing 200 μg/ml proteinase K (Sigma, UK) in 1 X PCR buffer (Sigma, UK). The material was incubated at 37°C for 24 hours and heated at 95°C for 15 minutes to deactivate the proteinase K. A total of 1 μl of the supernatant was used as template for DNA amplification.
DNA amplification A mastermix solution composed of 1 X Buffer II, 200 μM each dNTP (Promega, UK), 1.5 mM MgCl2, 1 unit Amplitaq Gold (Applied Biosystems), and 10 pmol each specific primer (MWG, Germany) was added to a reaction microtube (Table 2). A total of 1 μl of DNA was added to the mastermix. Cycling parameters were as follows: 95°C, 7 min, 1 cycle; 93°C, 45 s; 60°C, 45 s; 72°C, 1 min 30 s, 40 cycles; 72°C, 5 min, 1 cycle. For the FR3-trad protocol, the cycling parameters were as follows: 95°C, 7 min, 1 cycle; 93°C, 45 s; 55°C, 45 s; 72°C, 1 min 30 s, 40 cycles; 72°C, 5 min, 1 cycle.
Sequences of primers used in the different DNA amplification protocols
| Protocol and Size of Amplified Products | Primers-sequences (5′–3′) | |
|---|---|---|
| IgH FR2 Biomed Tube B 250 – 295 bp | VH1-FR2 | CTGGGTGCGACAGGCCCCTGGACAA |
| VH2-FR2 | TGGATCCGTCAGCCCCCAGGGAAGG | |
| VH3-FR2 | GGTCCGCCAGGCTCCAGGGAA | |
| VH4-FR2 | TGGATCCGCCAGCCCCCAGGGAAGG | |
| VH5-FR2 | GGGTGCGCCAGATGCCCGGGAAAGG | |
| VH6-FR2 | TGGATCAGGCAGTCCCCATCGAGAG | |
| VH7-FR2 | TTGGGTGCGACAGGCCCCTGGACAA | |
| JH | CTTACCTGAGGAGACGGTGACC | |
| IgH FR3 Biomed 100 – 170 bp | VH1-FR3 | TGGAGCTGAGCAGCCTGAGATCTGA |
| VH2-FR3 | CAATGACCAACATGGACCCTGTGGA | |
| VH3-FR3 | TCTGCAAATGAACAGCCTGAGAGCC | |
| VH4-FR3 | GAGCTCTGTGACCGCCGCGGACACG | |
| VH5-FR3 | CAGCACCGCCTACCTGCAGTGGAGC | |
| VH6-FR3 | GTTCTCCCTGCAGCTGAACTCTGTG | |
| VH7-FR3 | CAGCACGGCATATCTGCAGATCAG | |
| JH | CTTACCTGAGGAGACGGTGACC | |
| IgH FR3-trad 80 – 120 bp | FR3 | CCGAGGACACGGCCGTGTATTACTG |
| JH | AACTGCTGAGGAGACGGTGACC | |
| Ig kappa 130 – 150 bp | TUBE A | |
| VK1f/6 | TCAAGGTTCAGCGGCAGTGGATCTG | |
| VK2f | GGCCTCCATCTCCTGCAGGTCTAGTC | |
| VK3f | CCCAGGCTCCTCATCTATGATGCATCC | |
| VK4 | CAACTGCAAGTCCAGCCAGAGTGTTTT | |
| VK5 | CCTGCAAAGCCAGCCAAGACATTGAT | |
| VK7 | GACCGATTTCACCCTCACAATTAATCC | |
| JK1-4 | CTTACGTTTGATCTCCACCTTGGTCCC | |
| JK5 | CTTACGTTTAATCTCCAGTCGTGTCCC | |
| TUBE B | ||
| VK1f/6 | TCAAGGTTCAGCGGCAGTGGATCTG | |
| VK2f | GGCCTCCATCTCCTGCAGGTCTAGTC | |
| VK3f | CCCAGGCTCCTCATCTATGATGCATCC | |
| VK4 | CAACTGCAAGTCCAGCCAGAGTGTTTT | |
| VK5 | CCTGCAAAGCCAGCCAAGACATTGAT | |
| VK7 | GACCGATTTCACCCTCACAATTAATCC | |
| KDE | CCTCAGAGGTCAGAGCAGGTTGTCCTA | |
| INTR | CGTGGCACCGCGAGCTGTAGAC | |
| TCR-gamma tubes A and B 145–255 bp (A) and 80–220 bp (B) | Vγ1f (A) | GGAAGGCCCCACGCRTCTT |
| Vγ10 (A) | AGCATGGGTAAGACAAGCAA | |
| Vγ9 (B) | CGGCACTGTCAGAAAGGAATC | |
| Vγ11 (B) | CTTCCACTTCCACTTTGAAA | |
| JG11/21 | TTACCAGGCGAAGTTACTATGAGC | |
| JG13/23 | GTGTTGTTCCACTGCCAAAGAG | |
| Housekeeping genes – Biomed 100, 200, 300 and 400bp | TBXASI-US | GCCCGACATTCTGCAAGTCC |
| TBXASI-DS | GGTGTTGCCGGGAAGGGTT | |
| RAGI-US | TGAGCTGCAAGTTTGGCTGAA | |
| RAGI-DS | TGTTGACTCGATCCACCCCA | |
| PLFZ-US | TGCGATGTGGTCATCATGGTG | |
| PLFZ-DS | CGTGTCATTGTCGTCTGAGGC | |
| AF4-US | CCGCAGCAAGCAACGAACC | |
| AF4-DS | GCTTTCCTCTGGCGGCTCC | |
From: van Dongen JJ et al. Leukemia 2003 Dec;17(12):2257–317 and Diss TC et al. Mol Pathol. 2002 Apr;55(2):98–101.
PCR Analysis The IgH, IgL kappa and TCR-gamma genes are shown in Table 2. A negative control (no template) and a positive control (B-cell lymphoma) were used in each reaction. Amplification of non-rearranging genes to yield products of 100–400 bp was performed as a DNA quality control for each sample.14
Electrophoresis The PCR products (8 μl) were run on 6% or 8% polyacrylamide gels. Electrophoresis was performed at 200 V for 40 minutes, gels were stained with 0.5 μg/mL ethidium bromide for 5 minutes and the DNA bands were documented by use of the Gel Documentation System 1000 (Bio-Rad).
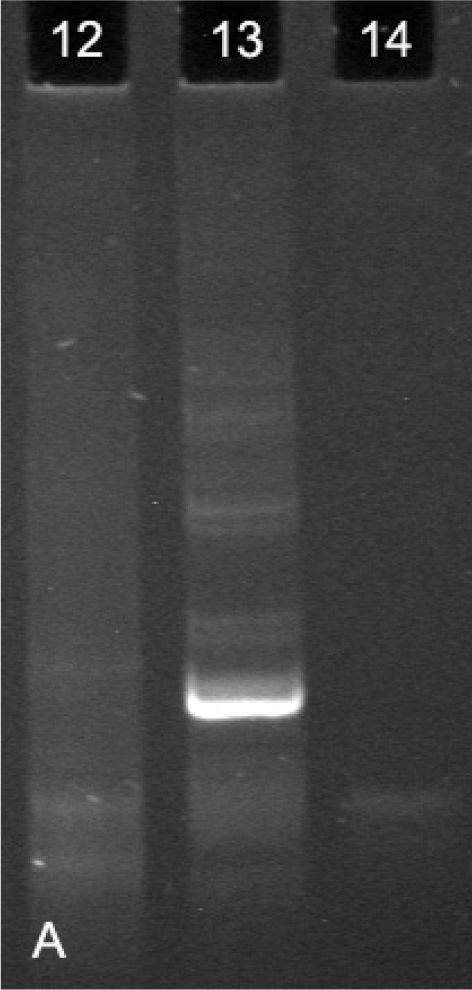
Evaluation of the Band Patterns The band patterns were analyzed, and each case was classified as follows: (i) Monoclonal (when one or two dominant bands of the same size were visualized in duplicate amplifications); (ii) Polyclonal (when a smear or variable ladder was detected); and (iii) Non-informative (when absence of bands or weak, non-reproducible patterns were observed) (Figure 1A).
Clonality analysis of a polyacrylamide gel following electrophoresis of PCR products using the IgL-K protocol. (12) Polyclonality (broad band, similar to a smear), (13) Monoclonality (well-defined, dominant band) and (14) Not defined (absence of bands or bands without definition)
A case was defined as monoclonal when at least one of the reactions showed a monoclonal pattern, independent of the results obtained from other reactions with different primers. The reactions were repeated separately or in duplicate (two identical reactions using two independent samples of DNA from the same case) for result confirmation in each case (Figure 2).
Clonality analysis of a polyacrylamide gel following electrophoresis of PCR products using the IgH Nizet protocol. Lane 1: positive control; lanes 2, 3, 4 and 5: polyclonal results (cases 2 and 4 in duplicate); lanes 6 and 7: monoclonal results (case 13 in duplicate); and lanes 8 and 9: undefined results (case 17 in duplicate)
Overall, monoclonality was observed in 10 cases, polyclonality in 12, and 7 cases were considered non-informative (Tables 3 and 4). The DNA of all samples showed low to moderate quality. When a housekeeping gene was analyzed, 8 cases yielded bands of only 100 bp, 15 cases yielded bands of 100 and 200 bp, and 6 cases yielded bands of 100, 200 and 300 bp. Bands of 400 bp were not observed (Figure 1B, Table 3). TCR-gamma rearrangement analysis did not reveal evidence of T cell monoclonality in any of the 29 cases studied.
Summary of clonality analysis results of the 29 cases with B-cell lymphoproliferative processes
| HE+IHC | Samples | Monoclonal | Polyclonal | Non-informative |
|---|---|---|---|---|
| PCBCL | 13 | 8 | 2 | 3 |
| B-PSL | 6 | 0 | 4 | 2 |
| INC | 10 | 2 | 6 | 2 |
HE: hematoxylin-eosin; IHC: immunohistochemistry; PCBCL: primary cutaneous B-cell lymphoma; NI: non-informative; B-PSL: B-cell pseudolymphoma; INC: inconclusive.
Clonality analysis of the 29 cases with B-cell lymphoproliferative processes
| I | HE+IHC | Clonality | HE+IHC+Clon. | WHO-EORTC | DNA | IgH F3 Nizet | IgH F3 Biomed | IgH F2 | IgL kappa | TCR |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | PCBCL | Monoclonal | PCBCL | follicle center | + | ND | NI | NI | monoclonal | NI |
| 2 | INC | Monoclonal | PCBCL | follicle center | ++ | monoclonal | NI | NI | polyclonal | polyclonal |
| 3 | PCBCL | NI | PCBCL | diffuse large B-cell, leg type | ++ | NI | NI | NI | NI | NI |
| 4 | INC | polyclonal | B-PSL | NA | ++ | polyclonal | NI | NI | polyclonal | polyclonal |
| 5 | B-PSL | NI | B-PSL | NA | +++ | NI | NI | NI | NI | NI |
| 6 | INC | polyclonal | B-PSL | NA | ++ | polyclonal | NI | NI | polyclonal | polyclonal |
| 7 | PCBCL | monoclonal | PCBCL | follicle center | ++ | monoclonal | NI | NI | monoclonal | polyclonal |
| 8 | B-PSL | NI | B-PSL | NA | + | NI | NI | NI | NI | NI |
| 9 | PCBCL | monoclonal | PCBCL | marginal zone | ++ | monoclonal | NI | NI | monoclonal | polyclonal |
| 10 | INC | NI | INC | NA | + | NI | NI | NI | NI | NI |
| 11 | PCBCL | monoclonal | PCBCL | follicle center | +++ | polyclonal | polyclonal | NI | monoclonal | polyclonal |
| 12 | INC | polyclonal | B-PSL | NA | + | polyclonal | NI | NI | polyclonal | NI |
| 13 | PCBCL | monoclonal | PCBCL | follicle center | ++ | monoclonal | NI | NI | monoclonal | polyclonal |
| 14 | PCBCL | NI | PCBCL | follicle center | + | NI | NI | NI | NI | NI |
| 15 | PCBCL | NI | PCBCL | follicle center | ++ | NI | NI | NI | NI | NI |
| 16 | B-PSL | polyclonal | B-PSL | NA | +++ | polyclonal | NI | NI | polyclonal | NI |
| 17 | INC | polyclonal | B-PSL | NA | ++ | polyclonal | NI | NI | NI | polyclonal |
| 18 | B-PSL | polyclonal | B-PSL | NA | + | polyclonal | NI | NI | NI | polyclonal |
| 19 | PCBCL | monoclonal | PCBCL | follicle center | ++ | polyclonal | NI | NI | monoclonal | polyclonal |
| 20 | B-PSL | polyclonal | B-PSL | NA | ++ | polyclonal | polyclonal | NI | polyclonal | polyclonal |
| 21 | INC | polyclonal | B-PSL | NA | +++ | polyclonal | NI | NI | polyclonal | polyclonal |
| 22 | PCBCL | polyclonal | PCBCL | follicle center | ++ | polyclonal | NI | NI | polyclonal | polyclonal |
| 23 | INC | monoclonal | PCBCL | follicle center | +++ | monoclonal | monoclonal | NI | monoclonal | polyclonal |
| 24 | INC | polyclonal | B-PSL | NA | + | polyclonal | NI | NI | polyclonal | polyclonal |
| 25 | PCBCL | polyclonal | PCBCL | marginal zone | ++ | polyclonal | NI | NI | polyclonal | polyclonal |
| 26 | INC | NI | INC | NA | ++ | NI | NI | NI | NI | NI |
| 27 | PCBCL | monoclonal | PCBCL | follicle center | + | polyclonal | NI | NI | monoclonal | polyclonal |
| 28 | PCBCL | monoclonal | PCBCL | follicle center | ++ | polyclonal | NI | NI | monoclonal | polyclonal |
| 29 | B-PSL | polyclonal | B-PSL | NA | +++ | polyclonal | NI | NI | polyclonal | polyclonal |
I: identification number of the case; HE: hematoxylin-eosin; IHC: immunohistochemistry; Clon: Clonality; PCBCL: primary cutaneous B-cell lymphoma; INC: inconclusive; NI: non-informative; NA: not applicable; B-PSL: B-cell pseudolymphoma; DNA: quality control of the DNA in each sample; (−) absence of DNA in the sample; (+) presence of DNA with 100 bp; (++) presence of DNA with 100 and 200 bp; (+++) presence of DNA with 100, 200 and 300 bp; TCR: T-cell receptor; NR: number of reactions.
Analyses of the IgH gene rearrangements in all cases were non-informative using the Biomed IgH FR2 method, and only three cases could be evaluated by the Biomed FR3 protocol (monoclonality in one case that was considered inconclusive at HE and IHC, and polyclonality in two cases considered PCBCL and 1 B-PSL, respectively, at HE and IHC). The IgH FR3-trad was the most useful protocol as 21 cases were conclusively classified. Monoclonality was observed in five cases (3 PCBCL and 2 considered inconclusive at HE and IHC), and polyclonality was observed in 16 cases (6 PCBCL, 4 B-PSL and 6 considered inconclusive at HE and IHC) (Tables 3 and 4).
Analysis of clonality through investigation of IgL-K gene rearrangements yielded results in 20 cases. Monoclonality was detected in 9 cases (8 PCBCL and 1 considered inconclusive at HE and IHC) and polyclonality in 11 cases (2 PCBCL, 3 B-PSL and 6 considered inconclusive at HE and IHC) (Tables 3 and 4).
Among the 10 cases considered inconclusive by morphology and IHC, monoclonality was demonstrated in two cases (20%) by investigation of IgH and IgL kappa gene rearrangements (Tables 3 and 4). Despite the small number of PCBCL cases, it was possible to observe monoclonality in 7 out of 10 cases that had been classified as primary cutaneous follicle center lymphoma and in 1 out of 2 cases that had been classified as primary cutaneous marginal zone B-cell lymphoma, according to the WHO-EORTC classification. It was not possible to define clonality in the single case of cutaneous diffuse large B-cell lymphoma, leg-type, by any of the protocols used in the present report.
The differential diagnosis between PCBCL and B-PSL is relevant for an appropriate follow up and treatment of patients with B-cell lymphoproliferative processes. However, the differentiation between such entities is complex and challenging.24 Since, to date, there is no “gold-standard” method that could be autonomously performed for such differentiation, this present study evaluated the diagnostic contribution of clonality analysis by investigating IgH and IgL clonality as an ancillary method to diagnose PCBCL.
The importance of the histological and immunohistochemical evaluation in the determination of PCBCL is incontestable, even though many cases may remain without diagnosis regardless the use of several immunohistochemical markers.25,26 In our study, 10 cases were considered inconclusive by histology and immunohistochemistry, which demonstrates the necessity of additional ancillary diagnostic methods. New markers such as IRTA-1 and MUM 1, tissue microarrays, chromosomal translocation analysis and fluorescence in situ hybridization (FISH) have been used to increase the diagnostic accuracy of PCBCL.27–33
The analysis of clonality by PCR-based tests has played an important diagnostic role in PCBCL, but appropriate protocols must be selected and optimized to ensure the improved diagnostic sensitivity of these techniques. DNA quality in the samples is of crucial importance and skin biopsies are notoriously poor in this respect. The process of tissue fixation in formalin and paraffin embedding variably damages the DNA by fragmentation or other mechanisms, thus impairing the sensitivity of the test and making it difficult to amplify relatively large fragments.14,20,34 These observations could explain the low to moderate quality of DNA found in our samples and the difficulty in amplifying the DNA, especially using the IgH FR2 protocol. This protocol amplifies a product of 250–295 bp, requiring a non-fragmented DNA sample. The same observation was also highlighted by other groups.14,19,35
In the present study, we noticed higher sensitivity of the IgH FR3 Nizet protocol compared to others (IgH FR3 Biomed and Ig FR2 Biomed) when the rearrangement of IgH genes was investigated. This protocol corroborated the diagnosis of three of the PCBCL and four of the B-PSL cases. The present finding can also be explained by the smaller size of the target DNA. The size of the target product in the IgH FR3 Nizet is of 80–120 bp, compatible with the DNA size present in the samples, while IgH FR3 Biomed and FR2 Biomed amplify larger nucleotide fragments (100–170 bp and 250–295 bp), requiring well-preserved DNA.
The main clinical contribution of our molecular investigation was the classification of two out of ten cases that had been previously considered inconclusive by histological and immunohistochemical methods. After determination of monoclonality by IgH gene rearrangement, these cases could be diagnosed as PCBCL. Although “false-positive” clonality results in benign cutaneous lesions are described in the literature, those two cases presented infiltrated lesions on the back that were clinically very suggestive of PCBCL (cases 2 and 23). Thus, the clonality analysis is of extreme relevance, mainly when analyzed in the context of the, histological and immunohistochemical data 36
Of subsidiary relevance, but still important, investigation of the Ig kappa gene rearrangements supported the previous diagnosis of eight of the PCBCL and three of the B-PSL cases. Amongst all cases shown to be monoclonal in the present study, this result was obtained only by Ig kappa gene rearrangement in five cases. It was striking that those five cases had a polyclonal result using IgH protocols. This finding indicates the need for analysis of both Ig heavy and Ig kappa genes.11,19
The importance of a monoclonal result in the confirmation of malignancy when analyzing a lymphocytic infiltrate is well defined in the literature.10 It is also well established that a polyclonal result does not exclude a diagnosis of lymphoma due to a high false-negative rate.9 In cases in which a diagnosis of PCBCL was made by histological and immunohistochemical methods, a polyclonal PCR result could be explained by DNA amplification of benign reactive cells obscuring the products from the expanded clone in addition to the difficulty of identification of a well-defined band in the electrophoresis gel or by failure of primer annealing to the tumor’s rearranged Ig gene segment.9,12,19 Laser-microdissection could be used to select specific cells for the DNA extraction process, which may improve the sensitivity.37
Based upon these findings, in our experience, the two most efficient molecular methods in this study, IgH FR3-trad and Ig kappa Biomed protocols, for analysis of formalin–fixed, paraffin-embedded samples could facilitate a differential diagnosis between PCBCL and B-PSL. Finally, interpretation of the clonality analysis in a clinical, histological and immunohistochemical context can be critical to the diagnosis of PCBCL.