The aim of this study was to simultaneously monitoring cytomegalovirus and human herpesvirus 6 active infections using nested-polymerase chain reaction and, together with clinical findings, follow the clinical status of patients undergoing liver transplant.
INTRODUCTION:The human β-herpesviruses, including cytomegalovirus and human herpesvirus 6, are ubiquitous among human populations. Active infections of human herpesvirus 6 and cytomegalovirus are common after liver transplantation, possibly induced and facilitated by allograft rejection and immunosuppressive therapy. Both viruses affect the success of the transplant procedure.
METHODS:Thirty patients submitted to liver transplant at the Liver Transplant Unit, at the Gastro Center, State University of Campinas, SP, Brazil, were studied prospectively from six months to one year, nested-polymerase chain reaction for cytomegalovirus and human herpesvirus 6 DNA detections. Two or more consecutive positive nested-polymerase chain reaction were considered indicative of active infection.
RESULTS:Active infection by cytomegalovirus was detected in 13/30 (43.3%) patients, median time to first cytomegalovirus detection was 29 days after transplantation (range: 0-99 days). Active infection by human herpesvirus 6 was detected in 12/30 (40%) patients, median time to first human herpesvirus 6 detection was 23.5 days after transplantation (range: 0-273 days). The time-related appearance of each virus was not statistically different (p = 0.49). Rejection of the transplanted liver was observed in 16.7% (5/30) of the patients. The present analysis showed that human herpesvirus 6 and/or cytomegalovirus active infections were frequent in liver transplant recipients at our center.
CONCLUSIONS:Few patients remain free of betaherpesviruses after liver transplantation. Most patients presenting active infection with more than one virus were infected sequentially and not concurrently. Nested-polymerase chain reaction can be considered of limited value for clinically monitoring cytomegalovirus and human herpesvirus 6.
The human β-herpesviruses, including cytomegalovirus (CMV) and human herpesvirus 6 (HHV-6), are ubiquitous among human populations.1 In Brazil, serological prevalence surveys conducted in the North and Southeast regions show adult infections rates for HHV-6 and CMV of around 90% among the population studied (0-40 years of age), with the primary infection occurring during the first year of life.2–4 Both viral agents can cause several human diseases either as a consequence of reinfection or reactivation of latent infection.5–7 The reactivation of latent HHV-6 and CMV is common following liver transplantation. The reactivation may possibly be induced and facilitated by allograft rejection and immunosuppressive therapy.8,9 Both viruses affect the success of the transplant procedure. Viral infection is associated with several observable clinical findings: fever, neutropenia, central nervous system manifestations or other visceral involvements.10 In addition, HHV-6 viremia is an independent significant predictor of invasive fungal infections and is associated with late mortality in liver transplantation recipients.11 In addition, coinfection with the viral agents can lead to a higher frequency of transplant rejection.12,13 The diagnosis of recrudescence or new infection with CMV and HHV-6 is not easy. Although serological detection techniques are available, the diagnostic value of a positive result is limited by the high prevalence of infection in adults.2,3 A report of specific anti-CMV or HHV-6 IgM in the sera or a four-fold rise in IgG antibodies can be used as a diagnostic criterion, but such an assay has limited sensitivity. Moreover, the interpretation of serological results is complicated by the fact that both primary and secondary infections with other herpes viruses may be associated with a concurrent antibody response to HHV-6.6,14 Amplification techniques are also available for the diagnosis of both viral agents; however, the results obtained using these techniques can be controversial, as they are dependent on the method of PCR employed.15–17 Nested PCR (N-PCR) amplifies one DNA target-specific sequence and is divided into two stages. In the first stage, a pair of primers amplifies the specific sequence of DNA. In the second, a new primer set is used for an internal region of the previously amplified fragment. In this second stage, the DNA is maintained at a high concentration to prevent nonspecific annealing, thus making this technique more efficient and specific.18,19 The aim of this study was to simultaneously monitor CMV and HHV-6 active infections using nested PCR and, together with clinical findings gathered over periods of six months to one year, follow the clinical status of patients undergoing liver transplants. CMV and HHV-6 antigenemia were also compared with the nested PCR results.
PATIENTS AND METHODSPatients - Thirty patients submitted for liver transplantation at the Liver Transplant Unit, Gastro Center in the State University of Campinas, SP, Brazil were studied prospectively for a period of six months to one year using N-PCR for CMV and HHV-6 DNA detection. For basic immunosuppressive therapy, the patients received steroids, azathioprine and cyclosporine. Tacrolimus (FK 506) and mycophenolate mofetyl (MMF) were prescribed based on selected patient characteristics and specific protocol studies. High doses of methyl prednisone were used as an anti-rejection treatment. All liver transplant recipients received 200 mg of acyclovir every 12 h for 60 days for prophylaxis against Herpes simplex infection because of the high seroprevalence of this virus in the Brazilian population (data not reported). Each patient protocol was revised and checked for clinical findings associated with infection by CMV or HHV-6. All episodes of rejection were documented by liver biopsy. Routine prophylaxis for CMV infection was not used unless the donor was positive and the recipient serum negative for CMV. Ganciclovir was administered for two weeks for protection against CMV in patients with two or more consecutive positive PCR results.20 No patients received anti-HHV-6 therapy during this study. Informed consent was obtained from each enrolled patient. The study protocol was approved by the Ethics Committee of the School of Medical Science at the State University of Campinas. All clinical and laboratory records were reviewed for evidence of HHV-6 and CMV-attributable disease symptoms.
Collection of blood samples - Peripheral blood samples were drawn weekly in the first month post-transplant. At thirty to 90 days post-transplantation, blood samples were collected every two weeks. After 90 days, samples were collected monthly up to one year post-transplant.
Peripheral blood leukocyte DNA extraction – The procedures for isolating blood leukocytes and DNA extraction for CMV have been described previously.14,21 The DNA amount and purity were determined by optical density readings obtained using spectrophotometry and by reading the absorbance at a wavelength of 260 nm (1 OD = 50 ng). The final DNA concentration present in each sample was determined by the formula OD 260×50×factor of dilution = ng/μl.
Serum DNA extraction - DNA was extracted from 200 μL of serum using a phenol-chloroform protocol after treatment with lysis buffer containing SDS and overnight incubation with proteinase K at 65°C, followed by DNA precipitation with cold ethanol. The resulting DNA pellet was eluted in 50 μL of TE buffer (Tris EDTA).22
CMV Nested PCR - Five microliters of DNA extracted from PBL, as described above, was used in the nested PCR. The reaction mixture contained primers specific to CMV and was generated following a previously described protocol.15,16 To exclude false-negative results, the DNA samples were subjected to PCR with β-globin primers.23
HHV-6 Nested PCR– Nested PCR was carried out using 5 μL of DNA extracted from serum as described above. The primers and protocol used for HHV-6 nested PCR have been previously described.24
All amplifications were carried out on a Robocycler 40 (Stratagene, La Jolla, CA, USA). The nested PCR products were analyzed under UV light after electrophoresis in 2% agarose (Gibco-BRL) and staining of the gel with ethidium bromide. All nested PCR reactions were carried out in duplicate using a second fresh aliquot.
CMV pp65-antigenemia- Blood samples were collected in EDTA-containing tubes and transferred to the laboratory within 6 hours. Leukocytes were isolated by a dextran sedimentation method that was followed by erythrocyte lysis. The cell pellet was suspended in phosphate-buffered saline (PBS), and the polymorphonuclear leukocytes (PML) were then centrifuged to prepare cytospin slides (3×105 PML per slide). The slides were air-dried and fixed in formaldehyde before being immunostained with monoclonal antibodies (Iq Products, Netherlands) and incubated with a peroxidase-labeled anti-mouse secondary antibody (HRP, Biotest, Dreieich, Germany). The test was carried out in duplicate, and the results were expressed as the number of positive cells per 3×105 PML.25–27
HHV-6 specific antigens in peripheral blood mononuclear cells (PBMCs)- The presence of the HHV-6 specific antigens in the PBMC cytopreparations was demonstrated by indirect immunoperoxidase staining using monoclonal antibodies against both the A and B variants (MAB8533 and MAB8535, Chemicon, Inc., Temecula, CA). Normal mouse IgG was used as a negative control for non-specific binding. A peroxidase-conjugated anti-mouse antibody (Dako, Copenhagen, Denmark) and a peroxidase-conjugated goat anti-rabbit antibody (Zymed, San Francisco, CA) were used as the second and third antibodies. A 3-amino-9-ethyl carbazole solution containing hydrogen peroxide was used as a stain. Mayer's hematoxylin was used for counterstaining.28,29
Definitions- Active CMV infection was defined as two or more consecutive N-PCR positive results and/or one positive pp65-antigenemia.5,26,30–32 CMV disease infection was determined based on biopsy and symptoms consistent with CMV disease including fever, malaise, myalgia, anorexia and leucopenia accompanied by active CMV infection.5,30 Patients were considered to have CMV viral syndrome if they suffered from unexplained fever (>37.5°C) for at least 3 days, in combination with at least one of the following features: arthralgia, leukopenia (<3×109/l), thrombocytopenia (<150×109/l) and liver enzyme elevation (ALT>50 U/l).33 A patient was considered to have asymptomatic CMV infection when active CMV infection occurred without the signs, symptoms, or laboratory abnormalities described above. A patient was considered to have an active HHV-6 infection based on one N-PCR positive result from serum-extracted DNA.
Statistical analysis- Categorical variables were compared using a Fisher's exact test or a chi-squared test. The Mann-Whitney-U test was used to compare continuous variables. p<0.05 was considered statistically significant.
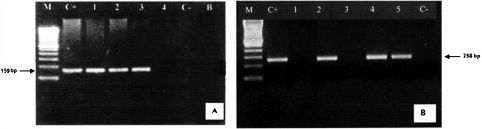
RESULTSThe nested PCR for CMV and HHV-6 was standardized with positive and negative DNA controls for each virus. Figure 1 illustrates the nested PCR products from CMV and HHV-6 positive and negative samples. Active infection of CMV was detected in 13/30 (43.3%) patients. The median time to initial CMV detection was 29 days following transplantation (range: 0-99 days). Active infection of HHV-6 was detected in 12/30 (40%) patients. The median time to initial HHV-6 detection was 23.5 days after transplantation (range: 0-273 days). The time to detection of each virus was not statistically different (p = 0.49). Co-infection did not occur frequently (Table 1), and CMV and/or HHV-6 DNA detection prior to positive CMV-pp65 antigenemia was observed in most patients.
Electrophoresis: A-N-PCR-amplified HCMV DNA of 4 peripheral blood samples (159 bp). B-N-PCR-amplified HHV-6 DNA of 5 peripheral blood samples. Agarose gel 2% ethidium bromide staining and photographed under UV light. M = molecular weight standard (Ladder 100 bp). C+ = positive control; C– = negative control; B = water.
Clinical and laboratory findings, day of onset and N-PCR for HCMV or HHV-6 in liver transplantation patients.
| Pt# | Clinical and Laboratory Findings | Onseta | Active Infections (aday) by N-PCR | Disease | ||
|---|---|---|---|---|---|---|
| HCMV | HHV-6 | Co-infection | ||||
| 1 | Absent | - | +(91) | - | - | No |
| 2 | Absent | - | +(66) | +(87) | + | No |
| 3b | Fever, granulocytopenia | 76 | +(181) | - | - | Yes |
| 4 | Granulocytopenia, thrombocytopenia | 19 | +(19) | - | - | Yes |
| 5b | Granulocytopenia, thrombocytopenia, diarrhea | 21 | +(20) | - | - | Yes |
| 6b | Fever, diarrhea | 93 | +(7) | +(300) | + | Yes |
| 7†.b | Fever, thrombocytopenia, granulocytopenia, | 127 | - | - | - | No |
| 8 | Granulocytopenia, thrombocytopenia, diarrhea, fever, mucosal ulcers | 30 | +(25) | - | - | Yes |
| 9 | Diarrhea, poliatralgia, myalgia, liver abscesses, colonic ulcers | 45 | - | - | - | No |
| 10 | Absent | - | +(18) | - | - | No |
| 11 | Fever, encephalitis | 24 | - | +(12) | - | Yes |
| 12 | Diarrhea | - | - | - | - | No |
| 13b | Diarrhea | 26 | - | - | - | No |
| 14† | Absent | - | - | +(98) | - | No |
| 15† | Fever, granulocytopenia | 84 | +(62) | +(62) | + | Yes |
| 16 | Absent | - | - | - | - | No |
| 17 | Absent | - | - | +(47) | - | No |
| 18 | Absent | - | - | +(6) | - | No |
| 19† | Absent | - | - | - | - | No |
| 20† | Absent | - | - | +(34) | - | No |
| 21† | Thrombocytopenia, encephalopathy, fever | 9 | - | +(12) | - | Yes |
| 22 | Absent | - | - | - | - | No |
| 23 | Absent | - | - | - | - | No |
| 24 | Fever, headache | 25 | +(26) | +(32) | + | Yes |
| 25† | Granulocytopenia, thrombocytopenia | - | - | +(77) | - | Yes |
| 26 | Granulocytopenia, thrombocytopenia | - | +(92) | - | - | Yes |
| 27 | Absent | - | +(114) | - | - | No |
| 28 | Thrombocytopenia, encephalopathy | 15 | +(51) | - | - | Yes |
| 29 | Absent | - | - | - | - | No |
| 30 | Absent | - | - | +(51) | - | No |
Pt# = patient numbers;
Overall, 53.3% (16/30) of patients presented viral infection-related symptoms and/or laboratory findings such as fever, granulocytopenia, thrombocytopenia, diarrhea, oral or intestinal ulcers and encephalitis (Table 1). Twenty percent (6/30) of patients had detectable CMV DNA and/or HHV-6 DNA in their serum associated with other laboratory and/or clinical manifestations of infection. Several cases demonstrated clinical manifestation, and CMV and/or HHV-6 active infection also were observed, but not concomitantly.
The rejection of the transplanted liver was observed in 16.7% (5/30) of the patients, although the time of the rejection episodes was not correlated to CMV and HHV-6 active infection (p>0.05) (Table 2). Most patients were successfully treated with steroids.
Patients who presented at least one episode of rejection during follow-up and episodes of CMV and/or HHV-6 active infection.
| Patient | CMV | HHV-6 | Grade of rejection |
|---|---|---|---|
| 3 | - | - | Mild (104) |
| 5 | +(20) | - | Moderate (493) |
| 6 | +(7) | +(300) | Mild (283) |
| 7 | - | - | Mild (105) |
| 13 | - | - | Moderate (85) |
∗ Numbers between parentheses represent day of onset.
The mortality rate at the one-year follow-up was 23% (7/30) (Table 3), and HHV-6 was detected in several cases. However, the deaths could not be correlated with active infections caused by either virus (p > 0.05).
Cause of patient death and episodes of CMV and/or HHV-6 active infection.
| Patient | CMV | HHV-6 | Cause of death |
|---|---|---|---|
| 7 | - | - | Gastrointestinal bleeding (178) |
| 14 | - | +(98) | Sepsis (293) |
| 15 | +(62) | +(62) | Sepsis (140) |
| 19 | - | - | Multiple organ failure (27) |
| 20 | - | +(34) | Sepsis (54) |
| 21 | - | +(12) | Graft failure (84) |
| 25 | - | +(77) | Base disease relapsed (98) |
∗ Numbers between parentheses represent day of onset.
The results of the present study indicate that HHV-6 and/or CMV infections were frequent in liver transplant recipients in the Gastro Center Hepatic Transplant Unit at the State University of Campinas (Campinas/Sao Paulo/Brazil). CMV and/or HHV-6 DNAemia was found in the majority of patients during the one year follow-up period. Our results are in agreement with the results of prior studies.9,34 These other studies suggest that the HHV-6 virus does not cause disease in itself but acts as a co-adjuvant to facilitate the emergence of several pathogens such as CMV.6,17,35 In the cohort studied, CMV and HHV-6 active infection data were in agreement with these reports. As there was no predominance of either type of viral infection, it was not possible to observe HHV-6 facilitation activity. The lack of predominance was probably because of the small sample number enrolled in this study (n = 30). Qualitative PCR, such as nested PCR, cannot distinguish between low and high viral load, and this may have contributed to the difficulty in determining significant viremia. However, in our center, N-PCR detected CMV DNA in two or more consecutive samples. These results had a considerable correlation with antigenemia testing (data not shown). N-PCR carried out with DNA extracted from serum is also frequently unable to detect latent infection.
Previous studies have noted the presence of CMV specific antigens in renal allograft biopsy material together with acute or chronic rejection, suggesting an association between viral infection and rejection.36 In the present study, this association could not be found because only one patient presented active infection by both agents, and HHV-6 DNAemia was not found during follow-up in any of the cases. In addition, the evaluation period of this study was not long enough to exclude the possibility that rejection may occur in patients co-infected after a time greater than one year. Active HHV-6 infection was observed in most of the patients who died after the one year follow-up, but this finding was not time-related.
CONCLUSIONThe results presented herein indicate that few patients remain free of β-herpesviruses following liver transplantation. Most patients with an active infection by more than one virus were infected sequentially and not concurrently. Active infections with HHV-6 or CMV might develop independently of each other. Most patients with HHV-6 active infections were asymptomatic, although several cases of HHV-6 active infection occurred among patients who died post-transplantation. Nested PCR is of limited value as a clinical CMV and HHV-6 monitoring tool.
All of the authors contributed substantially to the study. FAC designed the study, contributed to data analysis, performed the immunoassays and wrote the manuscript. PDA, SHAB and TCC contributed to data analysis and drafted the manuscript. AMS, MNS and RLT performed the immunoassays. CLR, IFB, ML, and LSL coordinated and participated in the study. SCBC contributed to the study design, conducted and coordinated the laboratory studies and wrote the manuscript. All authors read and approved the final manuscript.