The melanocortin receptor 4 (MC4R) participates in the control of appetite at the level of the central nervous system, through the leptin-melanocortin pathway. An association between different polymorphisms of the MC4R gene and obesity has been reported. However, there are few studies of the rs483145 single nucleotide polymorphism (SNP) of this gene.
ObjectiveTo investigate its prevalence and association with adiposity markers in Chilean adults.
MethodsThe prevalence of SNP rs483145, of the MC4R gene, was determined in 259 participants of the GENADIO study (genes, environment, diabetes and obesity) by means of real-time polymerase chain reaction (PCR). The association between the risk allele of MC4R (A) and adiposity markers (body weight, body mass index, fat mass percentage, hip circumference, waist circumference, waist-to-hip ratio) was performed by linear regression analysis and adjusted for confusion variables (socio-demographic and physic activity) using three statistical models.
ResultsIt was determined that the prevalence of the risk allele (A) of the SNP rs483145 of the MC4R gene is 24.5% in the Chilean adult population included in this study, without finding an association with any of the adiposity markers studied, both in adjusted and unadjusted models.
ConclusionThe presence of the risk allele of SNP rs483145 of the MC4R gene is not associated with adiposity markers in the Chilean adult population studied. New studies with a bigger sample size will be necessary to confirm these results.
El receptor de melanocortina 4 (MC4R) participa en el control del apetito a nivel del sistema nervioso central, a través de la vía de la leptina-melanocortina. Se ha reportado asociación entre diferentes polimorfismos del gen MC4R y la obesidad; sin embargo, existen escasos estudios del polimorfismo de nucleótido simple (SNP) rs483145 de este gen.
ObjetivoInvestigar su prevalencia y asociación con marcadores de adiposidad en adultos chilenos.
MétodosLa prevalencia del SNP rs483145, del gen MC4R, fue determinada en 259 participantes del estudio Genes, Ambiente, Diabetes y Obesidad (GENADIO) mediante reacción en cadena de la polimerasa (PCR) en tiempo real. La asociación del alelo de riesgo de MC4R (A) con marcadores de adiposidad (peso corporal, índice de masa corporal, porcentaje de masa grasa, perímetro de cadera, perímetro de cintura e índice cintura/cadera), se realizó mediante análisis de regresión lineal y fue ajustada por variables de confusión (sociodemográficas y de actividad física) mediante 3 modelos estadísticos.
ResultadosSe determinó que la prevalencia del alelo de riesgo (A) del SNP rs483145 del gen MC4R es del 24,5% en la población adulta chilena incluida en este estudio, sin encontrar asociación con ninguno de los marcadores de adiposidad estudiados, tanto en modelos ajustados como sin ajustar.
ConclusiónLa presencia del alelo de riesgo del SNP rs483145 del gen MC4R no se asocia con marcadores de adiposidad en la población adulta chilena estudiada. Nuevos estudios que incluyan una muestra más numerosa serán necesarios para confirmar estos resultados.
In recent years, obesity has become one of the most significant public health problems worldwide, accounting for more than 2.6 million deaths per year.1,2 It was initially recognised only as a risk factor for ischaemic cardiomyopathy; later, however, it was linked to diseases such as diabetes mellitus type 2, hypertension and cancer.3 Currently, 34.4% of the Chilean population is obese, and 74.2% of the adult population is overweight.4
The development of obesity is multifactorial, and combines modifiable factors, such as nutrition and physical activity, with non-modifiable factors, such as age, sex and genetic factors.5,6 At present, more than 700 genetic markers associated with common obesity have been reported, including single-nucleotide polymorphisms (SNPs) associated with the FTO gene, as they show the greatest strength of association, and melanocortin-4 receptor (MC4R) SNPs, as they were among the first to be identified.7,8
MC4R is a G-protein–coupled receptor linked to appetite control through the hypothalamic leptin–melanocortin pathway.9 Activation of MC4R occurs through its binding to molecules derived from pro-opiomelanocortin (POMC), such as the α and β forms of melanocyte-stimulating hormone (αMSH and βMSH).10 Agouti-related peptide (AGRP) is another MC4R-binding peptide that blocks anorexigenic signalling by POMC derivatives.11 Consequently, research has been conducted on the potential use of some synthetic MC4R agonists as drugs to treat obesity, especially in syndromic obesity of monogenic origin.12 Although the first agonists had adverse cardiovascular effects,13 subsequent phase-3 clinical studies of setmelanotide demonstrated significant appetite reduction and weight loss in patients with POMC or leptin receptor (LEPR) deficiency, paving the way for clinical use.14,15
Elimination of MC4R through genetic modification in mice causes obesity associated with hyperphagia and lack of satiety, while in humans mutations with loss of function are associated with severe early-onset monogenic obesity.16,17 These MC4R-inactivating mutations cause increases in body weight by up to 7 kg in homozygous individuals, but show a prevalence below 0.1% in Caucasian populations.18 In addition, MC4R has other high-prevalence genetic variants corresponding to SNPs, some of which are associated with a modest increase in the risk of obesity.15MC4R SNPs are distributed throughout the entire gene, including rs483145 and rs11872992 located in the promoter region, rs2229616 (V103I) and rs52820871 (I251 L) present in the only exon, and rs17782313 and rs12970134 located 188 kb and 150 kb from the 3′ end of the gene (distal locus), respectively.19 The latter SNPs are the most extensively studied and show an association with an increased risk of obesity in Asian, European and American people, reaching a variation by up to 0.3 kg/m2 in body mass index (BMI) per risk allele copy.20,21 By contrast, for the rs483145 SNP, just one study of association has been conducted, in a population of Pima Indians (indigenous people from Arizona, United States). This study found an increase in BMI by 0.58 kg/m2 per risk allele copy and a risk allele frequency over 80%.22 In the absence of studies linking the prevalence of the MC4R rs483145 SNP to obesity in the adult Chilean population, the objective of this study was to determine its prevalence in said population and to investigate its possible association with markers of adiposity.
Materials and methodsA cross-sectional descriptive study was conducted that included 259 individuals with available data for the genotype of the rs483145 SNP in the MC4R gene, belonging to the Genes, Ambiente, Diabetes y Obesidad [Genes, Environment, Diabetes and Obesity] (GENADIO) study, conducted in Chile from 2009 to 2011. The GENADIO project studied a total population of 475 individuals of Mapuche or European descent (249 and 226 individuals, respectively) from the Biobío and Los Ríos regions of Chile who had no history of metabolic or cardiovascular disease and who, at the time of the evaluation, had no prescription medications.23 The sample size was calculated based on CENSO 2002, the 2002 Chilean census, according to which indigenous people represented 4.6% of the country's population. To select participants of Mapuche or European descent and exclude individuals of mixed-race ancestry, only those whose paternal and maternal surnames were both of Mapuche or European origin, respectively, were included. In addition, to select Mapuche individuals, only those whose blood group was O were included. Participants were recruited by means of open invitations to the community or through community organisations such as primary healthcare centres, schools and social clubs. The study was approved by the independent ethics committees of the Universidad de Chile [University of Chile], Universidad de Concepción [University of Concepción] and University of Glasgow, who adhered to the Declaration of Helsinki on studies in humans. All participants signed an informed consent form prior to data collection.
Determination of allelic variants of the MC4R geneTo genotype the rs483145 SNP of the MC4R gene, genomic DNA was obtained from peripheral leukocytes using the QIAamp® DNA Blood Midi Kit (QIAGEN, Ltd. UK). Allelic discrimination was performed by means of real-time polymerase chain reaction (PCR) in an ABI 7900 H T thermal cycler using TaqMan® probes (Applied Biosystems, Warrington, UK). All sample testing was performed in duplicate, with a rate of success of genotype determination of 98%.
Markers of adiposityAnthropometric evaluation was performed by a trained professional using standardised protocols.24 Body weight and height were measured using electronic scales (Tanita TBF-300A, United States) and a stadiometer (SECA Model A800, United States) with a precision of 100 g and 1 mm, respectively. Nutritional status was classified based on the cut-off points suggested by the World Health Organization (WHO): underweight: <18.5 kg/m2; normal weight: 18.5–24.9 kg/m2; overweight: 25.0–29.9 kg/m2 and obesity: ≥30.0 kg/m2. Waist circumference (WC) was measured using non-stretch metric measuring tape (SECA Model 201, United States). The values used to define central obesity were the following: WC ≥ 94 cm in men and ≥80 cm in women.25 Hip circumference was measured around the fullest part of the hip region, at approximately the height of the pubic symphysis. Waist-to-hip ratio, determined by dividing waist circumference by hip circumference, was used to measure abdominal fat. Body composition was determined by measuring four skinfolds (biceps, subscapular, suprailiac and triceps) using a Harpenden calliper (Cranlea & Company, UK), with a precision of 0.2 mm.23,26 The Durnin–Womersley formula was used to estimate body fat percentage.26
Sociodemographic variables and physical activitySociodemographic data (age, sex, area of residence, ethnicity and level of education) were collected through validated surveys.19 Levels of physical activity (PA) and sedentary time were estimated by physical activity accelerometry (ActiGraph GT1M, United States). PA intensity and energy expenditure were determined using Freedson's algorithm.27
Statistical analysisThe data characterising the population studied are presented in terms of mean and standard deviation (SD) for continuous variables and percentage for categorical variables. Genotype differences were determined using regression analysis for continuous variables and the χ2 test for categorical variables. To investigate the link between the rs483145 polymorphism of the MC4R gene and markers of obesity (body weight, BMI, WC, hip circumference, waist-to-hip ratio and body fat percentage), linear regression analysis was performed. In addition, analyses of interaction between MC4R and sex and between MC4R and ethnicity were performed in order to determine whether the link between MC4R and markers of adiposity were different for men and women or for the Mapuche and non-Mapuche population. Since no evidence of a significant interaction was found (sex: p = 0.369; ethnicity: p = 0.560), it was not necessary to stratify the analyses of association by sex or ethnicity.
The rs483145 SNP genotype of the MC4R gene was coded according to an additive genetic model, where: 0 = homozygous TT for the protective allele, 1 = heterozygous AT for the risk allele and 2 = homozygous AA for the risk allele.
All analyses were adjusted for confounding variables using three statistical models: Model 0: not adjusted; Model 1: adjusted for age, sex, ethnicity, level of education and area of residence (urban/rural); and Model 2: adjusted for Model 1, but also for physical activity. The STATA® SE v14 software programme was used for all analyses. The level of significance was set at p < 0.05.
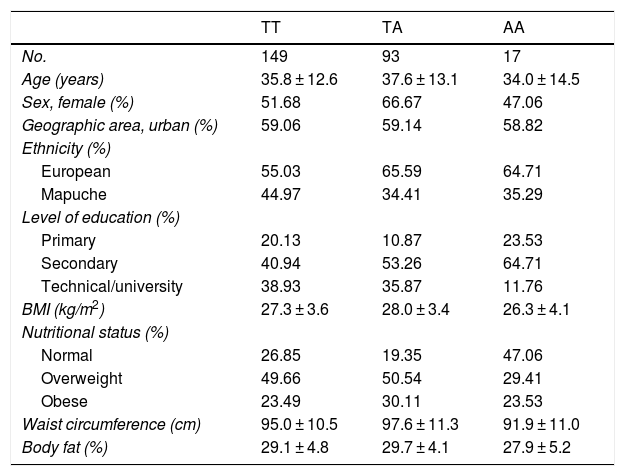
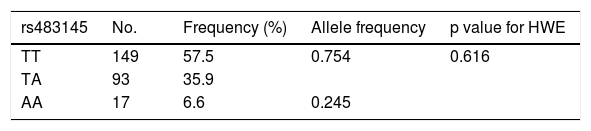
ResultsThe general characteristics of the study population by genotype are found in Table 1. The three genotypes had a similar average age: 35.8 for TT, 37.6 for TA and 34.0 for AA. It was found that 66.6% of those with the TA genotype, 51.6% of those with the TT genotype and 47.0% of those with the AA genotype were female. Lower rates of Mapuche ethnicity were seen among individuals with risk genotypes (34.4% for TA and 35.2% for AA) versus carriers of the protective TT genotype (44.9%). No appreciable between-group differences were seen in other sociodemographic variables or in parameters of physical activity. Table 2 shows the allele frequencies of the rs483145 SNP, distributed according to the Hardy–Weinberg equilibrium (χ2 = 0.616) and corresponding to 0.754 for the protective allele (T) and 0.245 for the risk allele (A).
Population characteristics by genotype of the MC4R gene (rs483145).
| TT | TA | AA | |
|---|---|---|---|
| No. | 149 | 93 | 17 |
| Age (years) | 35.8 ± 12.6 | 37.6 ± 13.1 | 34.0 ± 14.5 |
| Sex, female (%) | 51.68 | 66.67 | 47.06 |
| Geographic area, urban (%) | 59.06 | 59.14 | 58.82 |
| Ethnicity (%) | |||
| European | 55.03 | 65.59 | 64.71 |
| Mapuche | 44.97 | 34.41 | 35.29 |
| Level of education (%) | |||
| Primary | 20.13 | 10.87 | 23.53 |
| Secondary | 40.94 | 53.26 | 64.71 |
| Technical/university | 38.93 | 35.87 | 11.76 |
| BMI (kg/m2) | 27.3 ± 3.6 | 28.0 ± 3.4 | 26.3 ± 4.1 |
| Nutritional status (%) | |||
| Normal | 26.85 | 19.35 | 47.06 |
| Overweight | 49.66 | 50.54 | 29.41 |
| Obese | 23.49 | 30.11 | 23.53 |
| Waist circumference (cm) | 95.0 ± 10.5 | 97.6 ± 11.3 | 91.9 ± 11.0 |
| Body fat (%) | 29.1 ± 4.8 | 29.7 ± 4.1 | 27.9 ± 5.2 |
Data reported in terms of mean and standard deviation (SD) for continuous variables and percentage (%) for categorical variables.
BMI: body mass index; PA: physical activity.
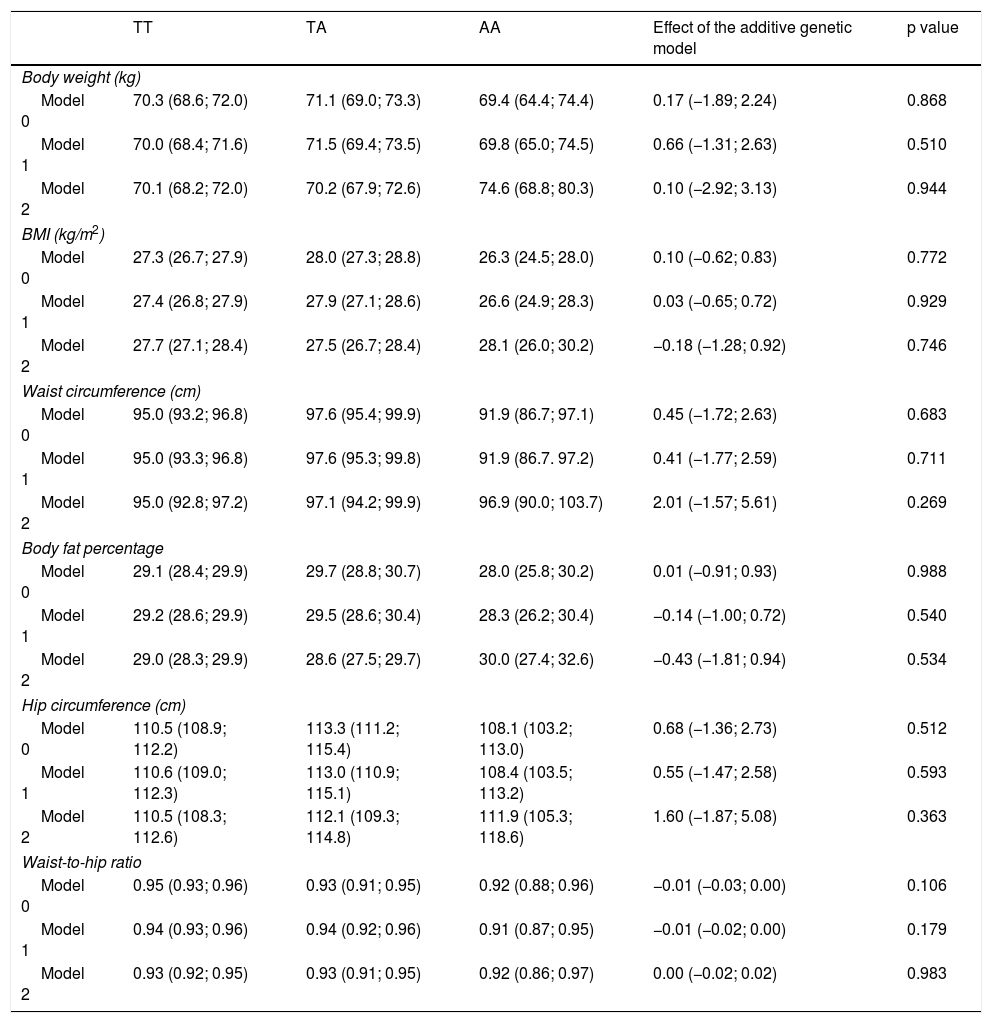
The results for the association between the rs483145 SNP of the MC4R gene and markers of adiposity are presented in Table 3. No significant associations were identified between the risk genotype and the markers of adiposity studied (body weight, BMI, waist circumference, body fat percentage, hip circumference and waist-to-hip ratio). For body weight, values of 70.3 kg for the protective TT genotype, 71.1 kg for the TA genotype and 69.4 kg for the AA genotype were seen in the non-adjusted model (Model 0). For all other markers of adiposity, apart from waist-to-hip ratio, similar variations in increases were seen for the TA genotype and similar variations in decreases were seen for the AA genotype, in Model 0 and Model 1, but they were not statistically significant.
Association between the genotype of the MC4R gene (rs483145) and markers of obesity.
| TT | TA | AA | Effect of the additive genetic model | p value | |
|---|---|---|---|---|---|
| Body weight (kg) | |||||
| Model 0 | 70.3 (68.6; 72.0) | 71.1 (69.0; 73.3) | 69.4 (64.4; 74.4) | 0.17 (−1.89; 2.24) | 0.868 |
| Model 1 | 70.0 (68.4; 71.6) | 71.5 (69.4; 73.5) | 69.8 (65.0; 74.5) | 0.66 (−1.31; 2.63) | 0.510 |
| Model 2 | 70.1 (68.2; 72.0) | 70.2 (67.9; 72.6) | 74.6 (68.8; 80.3) | 0.10 (−2.92; 3.13) | 0.944 |
| BMI (kg/m2) | |||||
| Model 0 | 27.3 (26.7; 27.9) | 28.0 (27.3; 28.8) | 26.3 (24.5; 28.0) | 0.10 (−0.62; 0.83) | 0.772 |
| Model 1 | 27.4 (26.8; 27.9) | 27.9 (27.1; 28.6) | 26.6 (24.9; 28.3) | 0.03 (−0.65; 0.72) | 0.929 |
| Model 2 | 27.7 (27.1; 28.4) | 27.5 (26.7; 28.4) | 28.1 (26.0; 30.2) | −0.18 (−1.28; 0.92) | 0.746 |
| Waist circumference (cm) | |||||
| Model 0 | 95.0 (93.2; 96.8) | 97.6 (95.4; 99.9) | 91.9 (86.7; 97.1) | 0.45 (−1.72; 2.63) | 0.683 |
| Model 1 | 95.0 (93.3; 96.8) | 97.6 (95.3; 99.8) | 91.9 (86.7. 97.2) | 0.41 (−1.77; 2.59) | 0.711 |
| Model 2 | 95.0 (92.8; 97.2) | 97.1 (94.2; 99.9) | 96.9 (90.0; 103.7) | 2.01 (−1.57; 5.61) | 0.269 |
| Body fat percentage | |||||
| Model 0 | 29.1 (28.4; 29.9) | 29.7 (28.8; 30.7) | 28.0 (25.8; 30.2) | 0.01 (−0.91; 0.93) | 0.988 |
| Model 1 | 29.2 (28.6; 29.9) | 29.5 (28.6; 30.4) | 28.3 (26.2; 30.4) | −0.14 (−1.00; 0.72) | 0.540 |
| Model 2 | 29.0 (28.3; 29.9) | 28.6 (27.5; 29.7) | 30.0 (27.4; 32.6) | −0.43 (−1.81; 0.94) | 0.534 |
| Hip circumference (cm) | |||||
| Model 0 | 110.5 (108.9; 112.2) | 113.3 (111.2; 115.4) | 108.1 (103.2; 113.0) | 0.68 (−1.36; 2.73) | 0.512 |
| Model 1 | 110.6 (109.0; 112.3) | 113.0 (110.9; 115.1) | 108.4 (103.5; 113.2) | 0.55 (−1.47; 2.58) | 0.593 |
| Model 2 | 110.5 (108.3; 112.6) | 112.1 (109.3; 114.8) | 111.9 (105.3; 118.6) | 1.60 (−1.87; 5.08) | 0.363 |
| Waist-to-hip ratio | |||||
| Model 0 | 0.95 (0.93; 0.96) | 0.93 (0.91; 0.95) | 0.92 (0.88; 0.96) | −0.01 (−0.03; 0.00) | 0.106 |
| Model 1 | 0.94 (0.93; 0.96) | 0.94 (0.92; 0.96) | 0.91 (0.87; 0.95) | −0.01 (−0.02; 0.00) | 0.179 |
| Model 2 | 0.93 (0.92; 0.95) | 0.93 (0.91; 0.95) | 0.92 (0.86; 0.97) | 0.00 (−0.02; 0.02) | 0.983 |
Data reported in terms of mean and 95% CI by genotype. The additive genetic model indicates the average increase in the adiposity variable for each additional copy of the risk variant (A). This additive effect and its respective 95% CI were determined by means of linear regression. The analyses were adjusted as follows: Model 0: not adjusted; Model 1: adjusted by age, sex, ethnicity, level of education, income, socioeconomic status and area of residence (urban/rural); and Model 2: adjusted for Model 1, but also for physical activity.
95% CI: 95% confidence interval; BMI: body mass index.
This study shows the absence of a significant association between the rs483145 SNP of the MC4R gene and markers of obesity in the adult Chilean population. The frequency of the risk allele (A) was estimated at 24.5%; this value was similar to that reported in European databases as 1000 genomes, but considerably lower than that of Pima Indians, who had a frequency of 80%.22 In addition, the rs17782313 SNP of the MC4R gene, which is more prevalent in the Caucasian population (30%), had a frequency of just 3% in Pima Indians, indicating that MC4R genetic variation depends heavily on ethnicity. In contrast with our results, the rs483145 risk allele in Pima Indians was associated with an increase by 0.58 kg/m2 in BMI per extra risk allele copy, although it was not associated with abnormalities in other markers of adiposity, such as body fat percentage or energy expenditure.22 The discrepancy in the association between the rs483145 SNP and BMI in Chileans versus Pima Indians may be linked to particular ethnic characteristics or to specific interactions with environmental factors that differ in these two populations, as demonstrated by longitudinal studies in which interactions between genetic make-up and environmental factors affected the degree of obesity present.28,29 However, the possibility that the lack of association was due to low statistical power cannot be ruled out, since the study conducted in Pima Indians included 11,268 participants compared to the 259 individuals included in this study.
Prior studies have examined the relationship between MC4R and obesity in the Chinese population, but by means of analysis of the rs17782313 SNP, a polymorphism located at the distal locus, 317 kb from the SNP studied herein.23 The rs17782313 SNP has a prevalence of the risk allele of nearly 30% in Chilean children and adults; it does not show a higher rate in individuals with obesity.30–32 Interestingly, these studies also revealed that, in both children and adults with obesity, the rs17782313 risk allele was correlated with abnormal eating behaviours, such as decreased levels of satiety and greater enjoyment of meals, but did not manage to determine any association with BMI.30 Taken together, studies of different genetic variants of MC4R conducted in the Chinese population have consistently found a high prevalence of risk variants close to 30% in various regions of the gene, with no association with obesity, possibly due to limited sample sizes (<500). In Europeans, on the other hand, the presence of the rs17782313 risk allele was accompanied by an increase by 0.2 kg/m2 in BMI, 0.66 kg in body weight and 14% in the likelihood of being obese.8 In the case of rs12970134, another distal-locus SNP, genome-wide association studies (GWAS) in Asian and European populations revealed an increase by 1 cm in waist circumference per risk allele copy.8
For SNPs associated with changes in the MC4R protein sequence, such as rs2229616 (V103I) and rs52820871 (I251 L), meta-analysis studies with more than 10,000 participants revealed that the minor alleles reduce the risk of obesity by 20% and 50% and have a frequency of 2%–7% in different populations, including American, Asian and European populations.3,8 For both SNPs, a decrease in BMI by close to 0.8 kg/m2 per risk allele copy has been reported. This effect is associated with a decreased response on the part of the MC4R gene to AGRP-mediated orexigenic signalling.19 Studies in Europeans also revealed abnormalities in food behaviour associated with non-synonymous variants and polymorphisms of the MC4R gene, such as a preference for fat consumption, lower sucrose intake and decreased response to satiety.33,34 The different studies reviewed revealed that the association between obesity and polymorphisms in MC4R and nearby loci are due to changes in the hypothalamic axis of appetite control, associated with response to POMC derivatives or AGRP, but also with regulation of macronutrient consumption in the amygdala, and with thermal homeostasis, associated with oxygen consumption in adipose tissue.9,34,35 Although the genetic variant analysed in this study is not located in the coding sequence, and therefore does not involve direct changes in MC4R protein function, its location towards the 5′ end of the gene renders it a potential cis-regulatory element with a potential impact on levels of MC4R expression. Should this be proven through, for example, expression quantitative trait loci (eQTL) studies, it might be postulated that carriers of the rs483145 SNP of MC4R could benefit from treatment with MC4R agonists which might increase signalling decreased by the effects of potentially lower receptor expression. Recently, the United States Food and Drug Administration (FDA) approved setmelanotide to treat severe obesity in patients with a POMC synthesis or leptin receptor deficiency, rendering it the first MC4R agonist to be approved for pharmacological use.
The limitations of this study included its limited sample size and the fact that it selected a population with an average age under 40 with no history of metabolic diseases. This study's limited sample size precluded analyses comparing men to women (112 and 147) or individuals of Mapuche descent to individuals of European descent (153 and 106). However, a lower prevalence of the risk allele could be seen in the Mapuche population (35% versus 65%). It should be noted that, in previously published studies in the same cohort, we identified positive associations between, on the one hand, genetic variants of FTO, TCF7 and SLC16A11 and, on the other hand, markers of obesity.36–38 The above suggests that any hidden association between the rs483145 SNP of the MC4R gene and obesity would have a significantly lower strength of association than those reported for FTO, TCF7 and SLC16A11. In addition, given that differences in food behaviours have been found for other SNPs of this gene, it would be useful to conduct similar studies for rs483145.16
ConclusionsOur results and the studies reported reveal the importance of ethnicity as a factor in the prevalence of the different SNPs of the MC4R gene, as well as the relationship thereof to markers of obesity. The different associations presented by other genetic variants of MC4R with obesity (predisposition versus protection) complexify the relationship between MC4R and obesity, revealing the need to study the prevalence of the various polymorphisms of MC4R in the different populations and their relationship to obesity, eating behaviours and metabolism. In particular, the rs483145 genetic variant was not found to be associated with obesity in the Chilean population, breaking with findings in Pima Indians.22 This study contributes new information that sheds light on the relationship between polymorphisms in the MC4R gene and obesity. This information could be taken into account in the design of primary prevention programmes and personalised treatment strategies in the Chilean population.
Conflicts of interestThe authors declare that they have no conflicts of interest.
FundingThis research has not received specific funding from public sector agencies, the commercial sector or non-profit organisations.
Please cite this article as: Mardones L, Parra-Valencia E, Petermann-Rocha F, Martínez-Sanguinetti MA, Leiva-Ordoñez AM, Lasserre-Laso N, et al. El polimorfismo rs483145 del gen MC4R no se asocia con obesidad en población chilena: resultados del estudio GENADIO. Endocrinol Diabetes Nutr. 2022;69:254–261.