Human papillomavirus (HPV) testing is increasingly used in cervical cancer prevention strategies, and a variety of HPV genotyping assays have been developed. We aimed to compare the performance of two HPV genotyping techniques in formalin-fixed paraffin-embedded (FFPE) tissue specimens from a series of invasive squamous cell carcinoma (SCC) cases.
MethodsArchival FFPE tissue blocks from 78 SCC cases were initially considered. DNA was extracted from dewaxed tissue sections and tested with the INNO-LiPA HPV Genotyping Extra assay (Innogenetics), and the F-HPV typing kit (Genomed) targeting the L1 and E6/E7 regions, respectively.
ResultsThe INNO-LiPA assay showed a higher sensitivity (98.6%) than the F-HPV assay (78.6%). A total of 12 (17.1%) biopsies showed multiple-type infections evidenced by at least one assay. Among the SCC cases tested, HPV16 and/or 18 were detected in 70% of the cases, and 18.4% of them had multiple infections with other high-risk types.
ConclusionsOur results suggest that the INNO-LiPA assay has a better performance than the F-HPV in FFPE specimens, probably due to its smaller amplicon size and the wider range of detectable HPV types. The prevalence of multiple infections could be higher than previously reported, as evidenced by the combination of the two assays.
La detección del virus del papiloma humano (VPH) es cada vez más utilizada en los algoritmos de prevención del cáncer cervical, y se ha desarrollado una gran variedad de ensayos para su detección y genotipado. Nuestro objetivo fue comparar dos técnicas de genotipado del VPH en muestras de tejido fijado y embebido en parafina (FFPE) derivadas de una serie de casos de carcinoma cervical invasivo de células escamosas (SCC).
MétodosSe incluyeron retrospectivamente 78 casos de SCC. La extracción de DNA se realizó después de cortar y desparafinar los tejidos FFPE. Se utilizaron los ensayos INNO-LiPA HPV Genotyping Extra assay (Innogenetics) y F-HPV typing kit (Genomed), los cuales amplifican las regiones L1 y E6/E7, respectivamente.
ResultadosEl ensayo INNO-LiPA mostró una sensibilidad más elevada (98.6%) que el F-HPV (78.6%). Un total de 12 (17.1%) biopsias mostraron infecciones múltiples, evidenciadas como mínimo por uno de los ensayos. Entre los casos de SCC evaluados, los tipos HPV16 y/o 18 fueron detectados en el 70% de los casos, y un 18.4% de ellos presentaron infecciones múltiples con otros tipos de alto riesgo.
ConclusionesNuestros resultados sugieren que el ensayo INNO-LiPA tiene un mayor rendimiento que el F-HPV en muestras FFPE, probablemente debido a su menor tamaño de amplicón y a un mayor número de tipos de VPH incluidos. La prevalencia de infecciones múltiples en el SCC podría ser mayor a la anteriormente reportada, tal y como indican los resultados combinados de las dos técnicas.
Invasive cervical cancer (ICC) is the third most common cancer in women worldwide, causing 250,000 deaths.1 In Spain, there are 18.83 million women at risk for developing ICC (15 years old or older).2 Having a persistent high-risk HPV genital infection is a necessary but not sufficient cause of ICC.3 About 40 different HPV types are known to infect the genital mucosa4; among them, 12 have been classified as group 1 carcinogens or high-risk types.5 Approximately 73% of all ICC cases worldwide are associated with either HPV16 or 18. Thus, HPV prophylactic type-specific vaccines based on HPV16 and 18 L1 virus-like particles have been developed and clinically tested,6,7 and are being administered in many countries around the world.8 Given that the distribution of the different HPV types varies geographically, population-based studies are important to assess the influence of local prevalence on vaccine efficacy for the prevention of cervical cancer. A variety of HPV genotyping assays have been developed (reviewed by M. Schiffman et al.),9 which target different HPV genomic regions, and offer the identification of a varying number of HPV types. Furthermore, other assay characteristics, such as the size of the amplified genomic region, may be relevant when using certain clinical specimens, such as formalin-fixed paraffin-embedded (FFPE) tissue.
The aim of this study was to compare the performance of two HPV genotyping techniques in FFPE tissue blocks from invasive squamous cell carcinoma (SCC) cases: the INNO-LiPA HPV Genotyping Extra assay (Innogenetics, Gent Belgium) targeting the capsid L1 gene, and the F-HPV typing kit (Genomed, Kemsing, United Kingdom) targeting the E6/E7 region with type-specific primers. We also determined the relevance of the assay used when determining the HPV type-specific prevalence, as well as the proportion of multiple infections.
MethodsPatients and specimensA total of 78 histopathologically confirmed invasive squamous cell carcinoma (SCC) cases were retrospectively included from HIV-negative women diagnosed between 1989 and 2010 at Hospital Universitari Germans Trias i Pujol, Badalona (Barcelona), which is a reference Centre that currently covers 800,000 inhabitants. The mean age at diagnosis was 52.6 years (standard deviation, 15.5). Archived formalin-fixed paraffin-embedded (FFPE) uterine cervical tissue specimens originally obtained for routine purposes were used and treated anonymously. This study was approved by the Ethics Committee at our institution.
FFPE tissue blocks, including biopsies, conisations, and uterine tissue from radical hysterectomy, were cut into 10μm-thick sections following strict conditions to avoid cross-contamination. Paraffin blocks containing non-HPV related cancers were also included and used as negative controls.
DNA extraction and HPV genotypingTissue sections were dewaxed with xylene (1mL) and washed twice with 100% ethanol. DNA was extracted from samples by using the NucleoSpin Tissue kit (Macherey-Nagel GmbH & Co. KG, Düren, Germany), following manufacturer's instructions. DNA concentrations were adjusted to 500ng DNA/PCR by spectrophotometric analysis using Nanodrop.
HPV DNA amplification was performed using INNO-LiPA HPV Genotyping Extra Amp (Innogenetics) kit, which uses consensus primers targeting a 65pb region in the HPV L1 gene. Genotyping was then performed by automated reverse hybridization with the INNO-LiPA HPV Genotyping Extra assay (Innogenetics), according to manufacturer's instructions. This assay is able to detect the following HPV types: HPV6, 11, 16, 18, 26, 31, 33, 35, 39, 40, 43, 44, 45, 51, 52, 53, 54, 56, 58, 59, 66, 68, 69, 70, 71, 73, 74 and 82 (it is not able to discriminate between HPV69 and 71). Specimens that were positive for HPV DNA but were not typeable with this assay were coded as HPVX (unknown genotype). This assay also amplifies a 280bp fragment of the human HLA-DPB1 gene to verify DNA quality and extraction efficiency, and uses uracil-N-glycosylase to avoid contamination by PCR products.
All samples were also tested with the F-HPV typing kit (Genomed), which uses specific primers to amplify fragments from 158 to 484bp in length in the E6/E7 region by multiplex fluorescent PCR, and detects 15 types (HPV6, 11, 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68), as well as a human STR used as internal control (140–180bp in length). Generated fragments were then analysed by capillary electrophoresis in an ABI PRISM 3130xl Genetic Analyzer (Applied Biosystems, Foster City, CA, USA).
Data analysisGiven that a larger number of HPV types are detected by the INNO-LiPA assay in comparison with the F-HPV assay, agreement between both genotyping methods was assessed, considering only those SCC cases that were positive by HPV types included in both assays. HPV and type-specific prevalences were expressed as percentages of cases positive by either assay out of all cases tested for HPV for which a valid result was obtained. The possible impact of HPV prophylactic vaccines was calculated assuming a 100% vaccine efficacy with and without cross-protection against related HPV types not included in the vaccine.
ResultsFive out of the 78 cases (6.4%) were not tested for HPV because of their inadequately low DNA concentration after extraction, mostly due to the small size of the tissue block. Among the 73 cases tested for HPV, four (5.5%) were negative by INNO-LiPA for both the human gene and HPV DNA. This number decreased to three (4.1%) after F-HPV type-specific PCR testing. These three negative results by both assays were excluded from the analysis, as they probably indicated either DNA degradation from the fixation procedure of FFPE specimens or PCR inhibition. While the specimens included in this study had been stored for varying periods of time until testing, these negative results were not associated to specimen storage time. Among the 70 samples that provided a valid result with either assay, 69 (98.6%) were positive by INNO-LiPA, 55 (78.6%) by the F-HPV assay (seven were HPV negative, and no HPV nor human DNA could be detected in eight), and 52 (74.3%) by both assays.
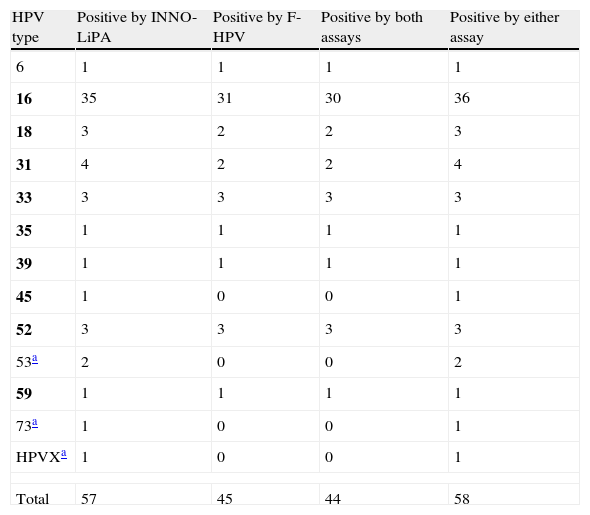
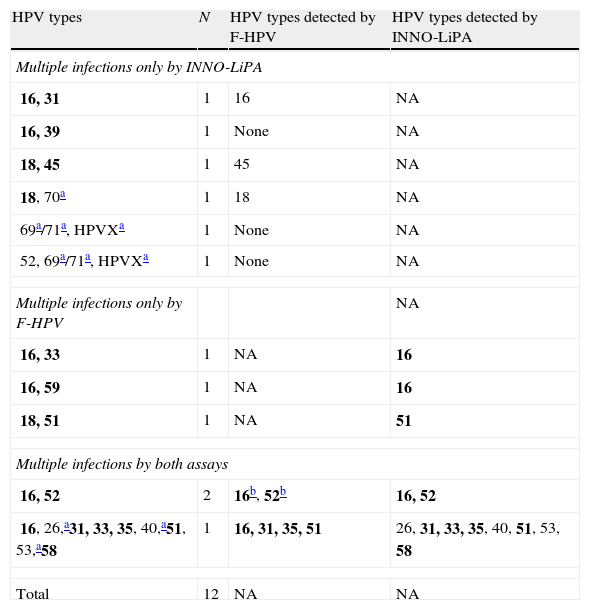
When the results of both assays were taken into account (i.e. a specimen was considered as positive for a given HPV type if positive by either one assay or the other), a total of 58 (82.9%) cases showed single-type infections (Table 1). Among them, an 81.5% agreement in HPV type calling was observed between both methods when only those HPV types included in both assays were taken into account (in 44 out of 54 cases, the same genotype call was obtained by both methods, nine cases were either negative or not detected by F-HPV, and one case was not detected by the INNO-LiPA assay). A total of 12 (17.1%) cases showed infection with multiple HPV types, with up to 6 HR types; this number varied between nine and six when considering the INNO-LiPA or the F-HPV assay, respectively. The agreement between assays was limited (Table 2).
Single HPV infections by both genotyping assays.
| HPV type | Positive by INNO-LiPA | Positive by F-HPV | Positive by both assays | Positive by either assay |
| 6 | 1 | 1 | 1 | 1 |
| 16 | 35 | 31 | 30 | 36 |
| 18 | 3 | 2 | 2 | 3 |
| 31 | 4 | 2 | 2 | 4 |
| 33 | 3 | 3 | 3 | 3 |
| 35 | 1 | 1 | 1 | 1 |
| 39 | 1 | 1 | 1 | 1 |
| 45 | 1 | 0 | 0 | 1 |
| 52 | 3 | 3 | 3 | 3 |
| 53a | 2 | 0 | 0 | 2 |
| 59 | 1 | 1 | 1 | 1 |
| 73a | 1 | 0 | 0 | 1 |
| HPVXa | 1 | 0 | 0 | 1 |
| Total | 57 | 45 | 44 | 58 |
HPV types included in the INNO-LiPA assay but not included in the F-HPV assay; HPVX, undetermined type. Group 1 carcinogenic HPV types according to the IARC classification5 are in bold.
Multiple HPV infections by either and both genotyping assays.
| HPV types | N | HPV types detected by F-HPV | HPV types detected by INNO-LiPA |
| Multiple infections only by INNO-LiPA | |||
| 16, 31 | 1 | 16 | NA |
| 16, 39 | 1 | None | NA |
| 18, 45 | 1 | 45 | NA |
| 18, 70a | 1 | 18 | NA |
| 69a/71a, HPVXa | 1 | None | NA |
| 52, 69a/71a, HPVXa | 1 | None | NA |
| Multiple infections only by F-HPV | NA | ||
| 16, 33 | 1 | NA | 16 |
| 16, 59 | 1 | NA | 16 |
| 18, 51 | 1 | NA | 51 |
| Multiple infections by both assays | |||
| 16, 52 | 2 | 16b, 52b | 16, 52 |
| 16, 26,a31, 33, 35, 40,a51, 53,a58 | 1 | 16, 31, 35, 51 | 26, 31, 33, 35, 40, 51, 53, 58 |
| Total | 12 | NA | NA |
NA, not applicable.
HPV types included in the INNO-LiPA assay but not included in the F-HPV assay; HPVX, undetermined type.
These types were detected in only one of the two cases. Types HPV69 and HPV71 cannot be distinguished by the INNO-LiPA assay. Group 1 carcinogenic HPV types according to the IARC classification5 are in bold.
By using the F-HPV assay, which targets the E6/E7 region, an additional HPV type was detected in four cases in comparison with INNO-LiPA (HPV types 16, 18, 33 and 59). On the other hand, HPV types detected by INNO-LiPA and included in the F-HPV assay were not detected on 18 occasions (HPV types 16, 18, 31, 45, 33, 39, 52 and 58).
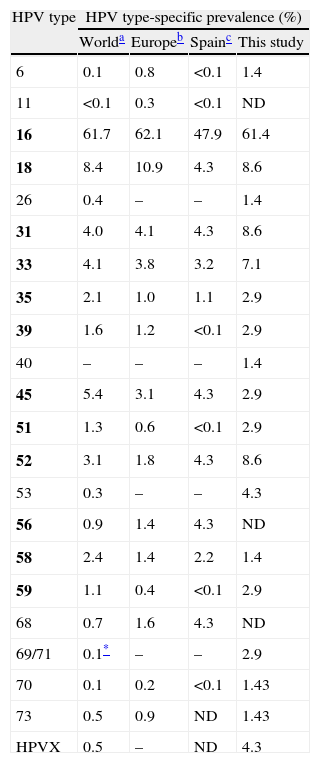
As the two assays were found to complement each other, their results were combined in order to estimate the HPV type-specific prevalence among the SCC cases studied. HPV16 was the most prevalent type, detected in 43 (61.4%) cases (Table 3). HPV18, 31 and 52 followed with 6 (8.6%) cases each. Overall, HPV16 and/or 18 were detected in 70% of the cases. However, seven (16.3%) of the 43 HPV16-infected women were also infected with other high-risk types, and this percentage was 50% for HPV52 and 33.3% for HPV18 and 31 (Table 2).
HPV type-specific prevalence in SCC cases in comparison with previous reports.
| HPV type | HPV type-specific prevalence (%) | |||
| Worlda | Europeb | Spainc | This study | |
| 6 | 0.1 | 0.8 | <0.1 | 1.4 |
| 11 | <0.1 | 0.3 | <0.1 | ND |
| 16 | 61.7 | 62.1 | 47.9 | 61.4 |
| 18 | 8.4 | 10.9 | 4.3 | 8.6 |
| 26 | 0.4 | – | – | 1.4 |
| 31 | 4.0 | 4.1 | 4.3 | 8.6 |
| 33 | 4.1 | 3.8 | 3.2 | 7.1 |
| 35 | 2.1 | 1.0 | 1.1 | 2.9 |
| 39 | 1.6 | 1.2 | <0.1 | 2.9 |
| 40 | – | – | – | 1.4 |
| 45 | 5.4 | 3.1 | 4.3 | 2.9 |
| 51 | 1.3 | 0.6 | <0.1 | 2.9 |
| 52 | 3.1 | 1.8 | 4.3 | 8.6 |
| 53 | 0.3 | – | – | 4.3 |
| 56 | 0.9 | 1.4 | 4.3 | ND |
| 58 | 2.4 | 1.4 | 2.2 | 1.4 |
| 59 | 1.1 | 0.4 | <0.1 | 2.9 |
| 68 | 0.7 | 1.6 | 4.3 | ND |
| 69/71 | 0.1* | – | – | 2.9 |
| 70 | 0.1 | 0.2 | <0.1 | 1.43 |
| 73 | 0.5 | 0.9 | ND | 1.43 |
| HPVX | 0.5 | – | ND | 4.3 |
WHO/ICO Information Centre on HPV and Cervical Cancer14 (based on a number of cases ranging between 493 and 3863 depending on the HPV type).
WHO/ICO Information Centre on HPV and Cervical Cancer2 (based on 188 cases for the first seven HPV types and on 46 for the rest).
Type HPV69; HPVX, undetermined type. Group 1 carcinogenic HPV types according to the IARC classification5 are in bold; HPV68 is classified as Group 2A probably carcinogenic; HPV26, 53, 69, 70, and 73 are classified as possibly carcinogenic types.
The low-risk HPV type HPV6 was detected in a single infection in one SCC case, as detected by both typing assays. Finally, non-typeable HPV types by INNO-LiPA (HPVX) were present in three cases (4.3%), one of them as a single infection, and the other two in mixed-type infections (Table 2).
DiscussionHPV genome integration is believed to be a key event in cervical carcinogenesis. Specifically, the HPV types most commonly associated with ICC worldwide (HPV16, 18 and 45) are found in the integrated state more frequently than other types (55–92%).10 Given that the L1 gene may be partially deleted during the integration process11,12 while the E6/E7 region is not affected, we compared the performance of two HPV genotyping assays targeting these two different genomic regions.
Employing the F-HPV assay, which uses HPV type-specific primers targeting the E6/E7 region, one additional case (HPV16) was detected as positive when compared with the INNO-LiPA assay targeting the L1 region, and one additional Group 1 carcinogenic type was detected in four cases. These results might be reflecting loss of the L1 region during integration. On the other hand, HPV types included in the F-HPV assay were not detected on 14 occasions. These results could be explained in FFPE specimens by the high degree of DNA degradation and cross-linking caused by the fixation process, which often leads to extracted DNA fragments of approximately 200bp or less in length.13 Thus, the smaller size of the amplicon obtained with the INNO-LiPA assay may have provided it with a higher sensitivity in this type of specimen as compared to the F-HPV assay, which amplifies larger regions of the HPV genome. Besides, 12 cases typed by INNO-LiPA as positive for HPV types included in the F-HPV assay were negative by the latter assay, which could reflect a higher sensitivity of the INNO-LiPA assay, or its higher robustness in the presence of PCR inhibitors (in 8 of those 12 cases no human DNA could be detected by F-HPV when using the same DNA eluate as for the INNO-LiPA assay). Lastly, the inclusion of a higher variety of HPV types in the INNO-LiPA assay also resulted in a higher percentage of HPV-positive cases (HPV types not included in the F-HPV assay were detected by the INNO-LiPA assay in five cases).
As the results of the two assays were considered to complement each other, they were combined in order to assess the HPV type-specific prevalence among the SCC cases studied. In agreement with the HPV16 prevalence observed in Europe (62.1–64.5%)14 and the world (62%),15 this type was also the most frequently associated with SCC in this study, with a higher contribution than previously reported in Spain (61.4% vs. 47.9% in 188 cases), as reviewed.2 HPV18, 31 and 52 followed in second place; HPV45 has been reported as the third and fourth most prevalent type in SCC worldwide15 and in Europe,16 respectively; while its prevalence in this study was somewhat lower. Conversely, HPV52 was as prevalent as HPV18 and 31 (8.6%), whereas it has been found in the sixth and seventh position worldwide and in Europe, respectively.15,16 Interestingly, in a previous study carried out in our centre on liquid cytology specimens from women with cytology results ranging from normal to HSIL, HPV52 was found in 4.4% of 160 HPV-positive HIV-negative women,17 suggesting that HPV52 prevalence is particularly higher in the SCC group. Although limited information regarding specific HPV type distribution in SCC is available in Spain,18–21 our data is in agreement with a review of previous studies,2 except for a somewhat lower prevalence of HPV45, and of HPV56 and 68, which were not found in any case.
HPV6 was detected in one case as a single infection. Similar results regarding this low-risk type have also been found in other studies; the contamination of biopsy specimens with surrounding lower-grade lesions has been suggested as a possible explanation for this fact, while the possibility remains that low-risk types can cause cancer in extremely rare circumstances.22–24 HPV70 was previously classified as a low-risk type, and HPV26 and 53 as probable high-risk types; recently, these three types have been classified as possibly carcinogenic.25 HPV70 and HPV26 were detected in mixed type infections with other high-risk types in two cases and, as reported in a previous study,26 HPV53 was the only type found in two SCC cases. This type showed a prevalence of 4.3% in this study, despite the small sample size, while this type is considered to be minor worldwide (<1% of all SCC cases).15,27
The two currently available HPV prophylactic vaccines offer protection against HPV16 and HPV18, the two most common oncogenic types worldwide (detected in 70–73% of all SCC cases).15,27 In agreement with global data, these vaccines would have prevented 49 (70%) of the cases diagnosed at our centre, if the prevalence takes into account both single and multiple infections, or up to a maximum of 57 cases (81.4%), if a 100% cross-protection is considered against related non-vaccine HPV types 31, 33 and 45. However, ten (20.4%) of those women infected by HPV16 and/or 18 had multiple infections with other HPV types, nine of them being Group 1 carcinogenic types. Thus, the fraction of women protected by these vaccines would go down to 39 (55.7%) if prevalence is considered in single infections only, or 50 (71%) if a 100% cross-protection is considered against related non-vaccine types. Although not all detected types may be relevant to a tumour, and HPV16 is regarded as a major carcinogen,28 the functional involvement of the other HPV types in multiple infections remains uncertain. While high-resolution HPV typing techniques have shown that each HPV type in a multiple infection in CIN biopsies could be associated with different morphologically discrete lesions,29 previous studies have found that cervical cancer patients with multiple infections showed a 5-fold increase in the treatment failure rate,30 as well as a shorter progression-free survival and cervical cancer-specific survival.31 Published prevalence studies usually report results obtained with a single genotyping assay; in fact, the observed prevalence of multiple infections in SCC in our study (17.1%), when combining the results of two assays was somewhat higher than that reported in recent worldwide reports (6.5–11.6%).15,27 The proportion of multiple infections observed may vary depending on a variety of factors of the specific genotyping method used, as demonstrated in this study, such as (i) the number of HPV types included in the genotyping assay; (ii) the sensitivity in the detection of each type; and (iii) the capacity of readily detected mixed infections. The detection of infections with two types belonging to the same α species (i.e. types 16 and 31 belong to phylogenetic group α-9) could be interpreted as an assay's cross-reactivity. In fact, a recent study showed that the tendency of infections with closely related HPV types to cluster together depended on the genotyping assay used.32 However, the reverse hybridization to type-specific probes in the LiPA assay takes place at high temperatures, and this assay has proven high specificity.33 Amplification is carried out in the F-HPV assay with type-specific primers that target the E6/E7 region, which is not as conserved as L1.
In conclusion, our results suggest that the INNO-LiPA assay has a better performance in HPV genotyping in FFPE specimens than the F-HPV assay. Even though this was a relatively small study, the combination of both assays provided us with more comprehensive results, and demonstrated that the prevalence of multiple infections involving several high-risk types might be higher than previously reported, which could influence the efficacy of available HPV prophylactic vaccines. Nevertheless, the cost-effectiveness of combining different genotyping methods should be carefully assessed depending on the goal of each study.
Conflict of interestsThe HPV genotyping reagents were partially provided by Innogenetics and Palex Medical SA (Sant Cugat del Vallès, Spain; distributor of Genomed). However, neither of the two commercial sponsors had an involvement in study design; collection, analysis, or interpretation of data; writing the manuscript; and the decision to submit the manuscript for publication.
This study was partially funded through internal support from the Microbiology Service, and grant number CP09/00044 (“Miguel Servet”) from “Ministerio de Ciencia e Innovacion” (MICINN), within the “Plan Nacional de Investigación Científica, Desarrollo e Innovación Tecnológica (I+D+I)” (EM). We would like to thank Elisabet Bascuñana for her help in sample processing.