Members of the genus Cryptosporidium are major contributors to the burden of diarrhoeal disease globally.1 Cryptosporidiosis primarily affect children in resource-poor settings with unsafe drinking water and inadequate sanitary facilities, but also represents a significant health concern in developed nations.2Cryptosporidium hominis and Cryptosporidium parvum, particularly the former, are the two major causative species of human cryptosporidiosis. To date, at least nine subtype families (Ia, Ib, Id, Ie, If, Ig, Ih, Ii, Ik) have been identified within C. hominis by sequence analysis of the 60-kDa glycoprotein (gp60) gene.3 Among them, Ia (1.7%), Ib (92.2%), Id (5.1%), and Ie (0.9%) were the subtype families found (n=529) in the four largest molecular epidemiological surveys conducted so far in Spain.4–7 In these studies, all the Ib samples (n=152) molecularly characterized at the sub-genotype level were assigned to IbA10G2.
In December 2017 an 18-month-old male infant complaining of gastrointestinal symptoms including altered intestinal transit, distended abdomen, cramps, and acute, non-bloody watery diarrhoea associated to weigh loss (below 25th percentile) and anaemia (serum iron: 24μg/dL) was admitted to the outpatient clinic of the University Hospital Puerta de Hierro Majadahonda (Madrid) for routine coproparasitological examination. The patient had a normal immune status, no contact with pet animals, and no relevant record of recent travelling abroad, although his father reported travelling to Romania during the same period. A single, concentrated stool sample tested positive for the presence of Cryptosporidium oocysts by a rapid immunochomatographic test (Cer Test Biotec S.L., Zaragoza, Spain) and by microscopic examination of a fresh faecal smear stained with the modified Ziehl–Neelsen method. As part of an ongoing research project, a new, fresh aliquot of the faecal material was sent to the National Centre for Microbiology at Majadahonda (Madrid) for genotyping analyses. Total DNA was extracted and purified using the QIAamp® DNA stool mini test kit (Qiagen, Hilden, Germany). Molecular characterization of the sample was achieved by PCR amplification of the gp60 marker.8 Amplicons of the expected size (∼870bp) were directly submitted for sequencing. Subsequent sequence analyses confirmed the presence of C. hominis 1bA12G3, a sub-genotype not previously identified in Spain. A representative nucleotide sequence of the sub-genotype obtained was submitted to the GenBank® public repository under the accession number MH161561.
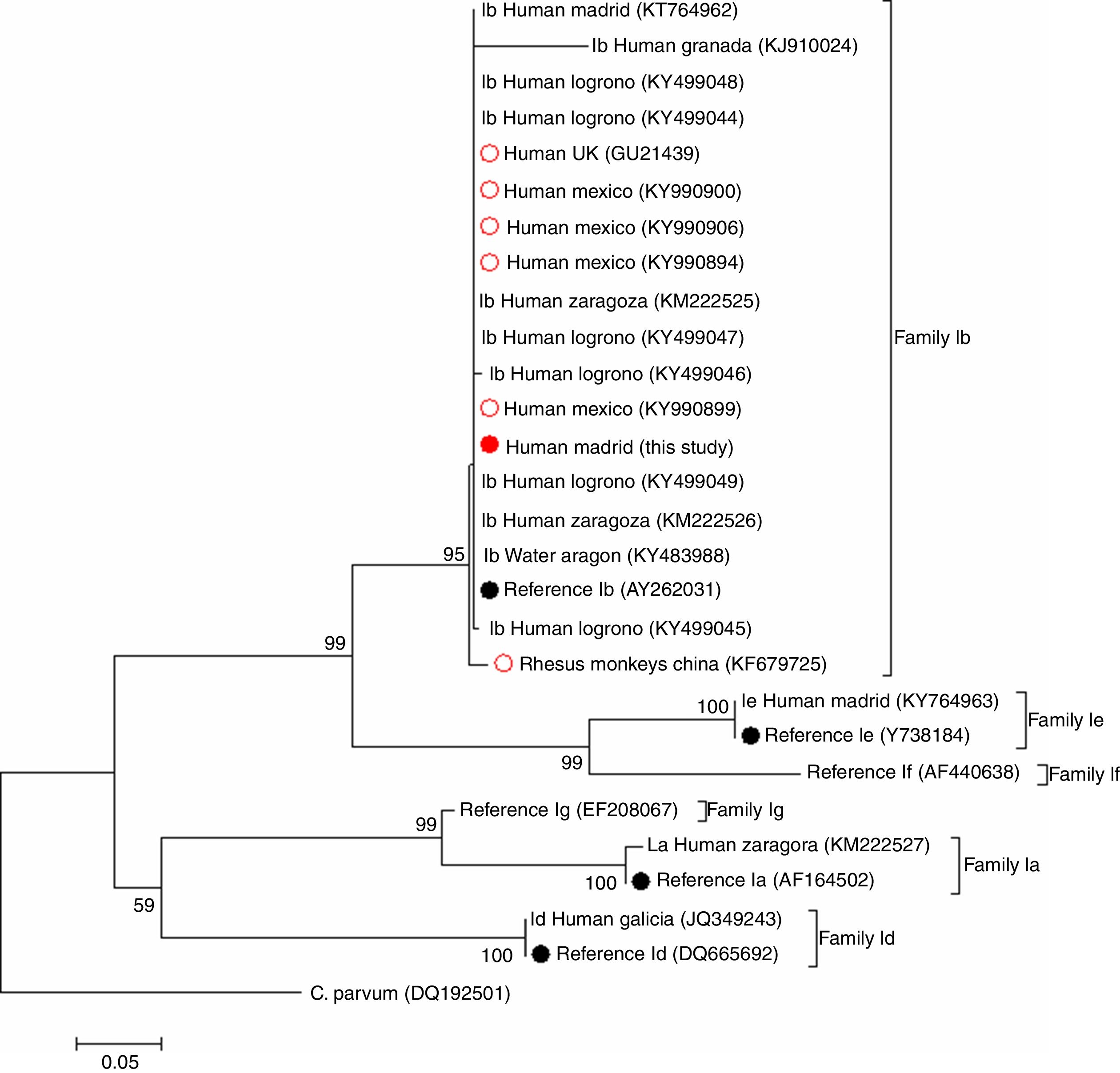
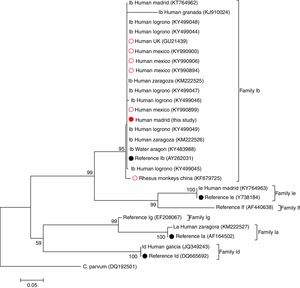
Remarkably, a search using the BLAST tool of the National Centre for Biotechnology Information (NCBI) revealed that only six additional IbA12G3 sequences have been previously deposited in GenBank®. This particular C. hominis sub-genotype was initially described in a human specimen from UK in 2010.9 Further molecular research conducted in the Sonora state (Mexico) allowed the identification of IbA12G3 in four children attending hospital settings, three of them presenting with gastrointestinal and/or nutritional disorders and the remaining one asymptomatic at the moment of diagnosis.10 Finally, IbA12G3 has also been reported in seven rhesus macaques (Macaca mulatta) housed on monkey farms in China, a finding that may be indicative of potential zoonotic transmission.11Fig. 1 shows the phylogenetic relationships among the IbA12G3 sequences generated in the present study and those reported in the surveys mentioned above. Appropriated reference sequences and representative sequences of the most frequent C. hominis family subtypes circulating in Spanish human populations were retrieved from NCBI and included in the analysis for comparative purposes. As expected, sequences belonging to the family subtype Ib grouped together in a well-defined cluster. Within this group, the IbA12G3 sequence of non-human primate origin was placed into an independent (but closely related) clade, very likely reflecting differences in host adaptation and specificity.
Phylogenetic tree illustrating the evolutionary relationship of the IbA12G3 sequence generated in the present study (represented by a red filled circle) to other C. hominis family Ib sequences previously identified in Spain and other countries at the GP60 locus inferred by a neighbour-joining analysis. Red empty circles represent all human (n=5) and non-human (n=1) primate IbA12G3 sequences reported globally to date. Black filled circles indicate reference sequences from GenBank®. Bootstrapping values over 50% from 1.000 replicates are shown at the branch points. The evolutionary distances were computed using the Tamura 3-parameter method. The rate variation among sites was modelled with a gamma distribution (shape parameter=1). C. parvum was used as outgroup taxa.
In summary, we show here the first description in Spain of a C. hominis human infection by IbA12G3, a very rare sub-genotype only reported before in clinical specimens in UK and Mexico. A host-adapted IbA12G3 variant has been previously identified infecting non-human primates in China, raising doubts about its actual zoonotic potential. Although the origin of the infection in our paediatric case could not be elucidated, we cannot rule out the possibility that this C. hominis sub-genotype is naturally circulating in Spain at low frequency rates. This study clearly demonstrates that molecular surveillance of human cryptosporidiosis is key in determining the occurrence or emergence of novel genotypes of the parasite of uncertain pathogenicity and virulence.
FundingThis study was funded by the Health Institute Carlos III (ISCIII), Ministry of Economy and Competitiveness under project MPY 1350/16.
The authors acknowledge Marta Hernández de Mingo and María Guerrero for excellent technical assistance, and Dr. Consuelo López for providing relevant epidemiological and clinical information.