Our objective was to characterize the enzymatic β-lactam resistance in clinical Enterobacteriaceae isolates with diminished susceptibility to carbapenems from 2013 to 2014 at Hospital Universitario Miguel Servet.
Material/methodsA total of 63 clinical isolates were analyzed for the presence of carbapenemases (KPC, OXA-48 and MBL), ESBLs and AmpC enzymes by combined disk methods and PCR detection of carbapenemase-encoding and beta-lactamase-encoding genes.
ResultsFifteen isolates had a phenotypic test compatible with carbapenemase production; two of these were confirmed by PCR as OXA-48 producers. ESBL detection was positive in 27 isolates (43%); plasmid-mediated AmpC was detected in nine isolates (14.2%) and derepressed AmpC β-lactamase was present in 18 isolates (28%).
ConclusionDuring the study period, the decreased susceptibility to carbapenems in Enterobacteriaceae in our area was not due to true carbapenemases but rather to β-lactamase activity (82.5% were ESBL or AmpC producers), probably in combination with decreased permeability of the outer membrane.
Nuestro objetivo fue caracterizar la resistencia enzimática a β-lactámicos en aislados clínicos de Enterobacteriaceae con sensibilidad disminuida a carbapenems desde 2013 a 2014 en el Hospital Universitario Miguel Servet.
Material/métodosSe analizaron un total de 63 aislados clínicos para presencia de carbapenemasas (KPC, OXA-48 y MBL), BLEE y AmpC por método de discos combinados y detección de genes codificantes de carbapenemasas y betalactamasas por PCR.
ResultadosQuince aislados tuvieron un test fenotípico compatible con producción de carbapenemasas; dos de ellos confirmados por PCR como productores de OXA-48. La detección BLEE fue positiva en 27 aislados (43%); se detectó AmpC plasmídica en 9 aislados (14,2%) y se observó β-lactamasa AmpC desreprimida en 18 aislados (28%).
ConclusiónDurante el período de estudio, la sensibilidad disminuida a carbapenems en Enterobacteriaceae en nuestra área no se debió a verdaderas carbapenemasas sino a actividad β-lactamasa (82,5% eran productores de BLEE o AmpC) probablemente en combinación con permeabilidad disminuida de la membrana externa.
Carbapenemase-producing Enterobacteriaceae (CPE) has become a public health concern and a threat for the treatment of patients infected with these organisms. Severe infections caused by these bacteria are associated with high mortality rates and rapid nosocomial and community spread, causing important outbreaks in hospitals worldwide.1 Therapeutic options for the treatment of these infections are limited as these bacteria often acquire a multi-drug resistance profile of difficult control.2
In addition, diagnosis and detection is a challenge for microbiology laboratories, since these enzymes do not always define resistance to carbapenems.1,3 A fast and early detection of carbapenemase-producers is essential to carry out successful strategies.
Carbapenem resistance can also be produced in the absence of carbapenemases through the derepression of the constitutive AmpC-encoding gene, the acquisition of exogenous plasmid-borne cephalosporinase- or extended spectrum β-lactamase (ESBL)-genes, or via porin permeability reduction.4 The aim of this study was to characterize the enzymatic mechanisms of resistance to β-lactams in clinical isolates of Enterobacteriaceae showing diminished susceptibility to carbapenems between January 2013 and December 2014 at our hospital.
Material and methodsSelection of bacterial isolates and inclusion criteriaDuring the study period, 88 clinical isolates of Enterobacteriaceae presented diminished susceptibility to carbapenems; among these, only 63 were available for this study. Antimicrobial susceptibility was performed by an automated broth microdilution method (Microscan Walkaway®, Beckman Coulter, Spain) and/or by the disk diffusion method. According to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoints for CPE screening and taking into consideration the MIC range of our commercial systems, we included isolates with imipenem MIC ≥1mg/l (Panel NC53 or Panel NM44, Microscan Walkaway®) or ≥2mg/l (Panel NM37, Microscan Walkaway®), meropenem MIC ≥1mg/l and/or ertapenem MIC ≥0.5mg/l.
The imipenem epidemiological cut-off value (ECOFF) according to EUCAST (MIC=4mg/l) was taken into account for isolates in which intrinsic decreased susceptibility to imipenem has been described (Proteus spp., Providencia spp. and Morganella spp.). Thus, within these genera, we only included isolates with imipenem MIC ≥8mg/l.
Phenotypic detection of ESBLs, AmpC enzymes and carbapenemasesAll isolates were screened for the presence of carbapenemases by combined disk diffusion methods (KPC, OXA-48 and metalobetalactamases (MBL)). Two tests were used depending on the isolation date. Until August 2013 the method contained a carbapenem (meropenem 10μg, Oxoid) alone and in combination with a KPC inhibitor (phenylboronic acid, PBA (Sigma–Aldrich) 400μg added), a MBL inhibitor (ethylenediaminetetraacetic acid, EDTA (Sigma–Aldrich), 292μg added) and meropenem-PBA-EDTA (400μg/292μg).5
For the interpretation of results, the inhibition zone of the carbapenem disk was compared to the inhibition zones of each of the carbapenem-plus-inhibitor disks. Therefore, production of KPC was suspected when a difference of ≥5mm was observed between meropenem plus PBA as well as meropenem-PBA-EDTA and meropenem. MBL production was suspected when the difference was ≥5mm between meropenem plus DPA as well as meropenem-PBA-EDTA and meropenem. If only a difference of ≥5mm was observed between meropenem-PBA-EDTA and meropenem but were no difference (<5mm) with the other carbapenem-plus-inhibitor disks, both KPC and MBL were then suspected.
From September 2013 we used the KPC/MBL and OXA-48 Confirm Kit (RoscoDiagnostica A/S, Taastrup, Denmark) as the routine method for screening of carbapenemases with the aim of detecting OXA-48 carbapenemase-producers and improve the sensitivity. The kit contains disks of meropenem 10μg, meropenem plus dipicolinic acid, DPA (MBL inhibitor), meropenem plus cloxacillin (AmpC inhibitor), meropenem plus PBA (KPC inhibitor) and temocillin 30μg (OXA-48-like screening).6 The tests were interpreted according to manufacturer's instructions as follows. If the meropenem plus DPA disk showed an inhibition zone ≥5mm than that of the meropenem disk, the organism was compatible with MBL activity. An inhibition zone difference ≥5mm between meropenem plus cloxacillin and meropenem alone indicated AmpC activity. If the inhibition zone difference was ≥4mm with meropenem plus PBA but there was no difference (<4mm) with the meropenem plus cloxacillin indicated the presence of a KPC enzyme. Negative results of all synergy tests and no zone of inhibition with temocillin 30μg were presumptive of an OXA-48 or OXA-48-like enzyme.
Isolates with decreased susceptibility to cephalosporins which fulfilled criteria according to EUCAST breakpoints for ESBLs and/or AmpC screening were phenotypically analyzed, by using combined disk methods with cefotaxime, ceftazidime and/or cefepime plus clavulanic acid7 for ESBLs detection and with cefotetan plus boronic acid for AmpC type beta-lactamases detection.8
A positive result was interpreted by a difference of ≥5mm in the inhibition zone in the presence of the inhibitor compared with that of the β-lactam alone.
PCR detection of β-lactamase genesAll isolates were analyzed for the presence of carbapenemases by a multiplex PCR assay, according to the method described by Poirel et al.9 Sets of primers for the following genes were included: blaNDM, blaVIM, blaKPC and blaOXA-48.
In Escherichia coli, Klebsiella spp. and Proteus spp. isolates with ESBLs and/or AmpC positive phenotypic tests, additional betalactamases encoding genes were investigated: blaSHV, blaTEM and blaCTX-M10 and six sets of AmpC-specific primers (ACC, FOX, CMY/MOX, DHA, LAT/CMY/BIL and ACT/MIR11). In chromosomal AmpC producers with phenotypic ESBL detection, blaSHV, blaTEM and blaCTX-M10 genes were also analyzed.
DNA extraction was performed using PrepMan® Ultra Sample Preparation Reagent (Applied Biosystems) according to the manufacturer's instructions. PCR products were visualized by electrophoresis in 2% agarose gels stained with GelRed™Nucleid Acid Stain 10,000 in water (Biotium).
ResultsA total of 88 non-duplicated clinical isolates of Enterobacteriaceae showed reduced susceptibility to carbapenems, 0.4% over 20.774 Enterobacteriaceae isolated during the study period; among the 88 isolates, 63 were available to be included in the study. The specimens were collected from 22 urine samples, 15 respiratory secretions, 10 wound samples and biological fluids, 9 blood cultures and 7 screening nasal, rectal and/or inguinal swabs for epidemiological surveillance. At the time of isolation, 55.5% of the patients (n=35) were hospitalized, 38% were outpatients (n=24) and 6.3% (n=4) were admitted at the emergency department.
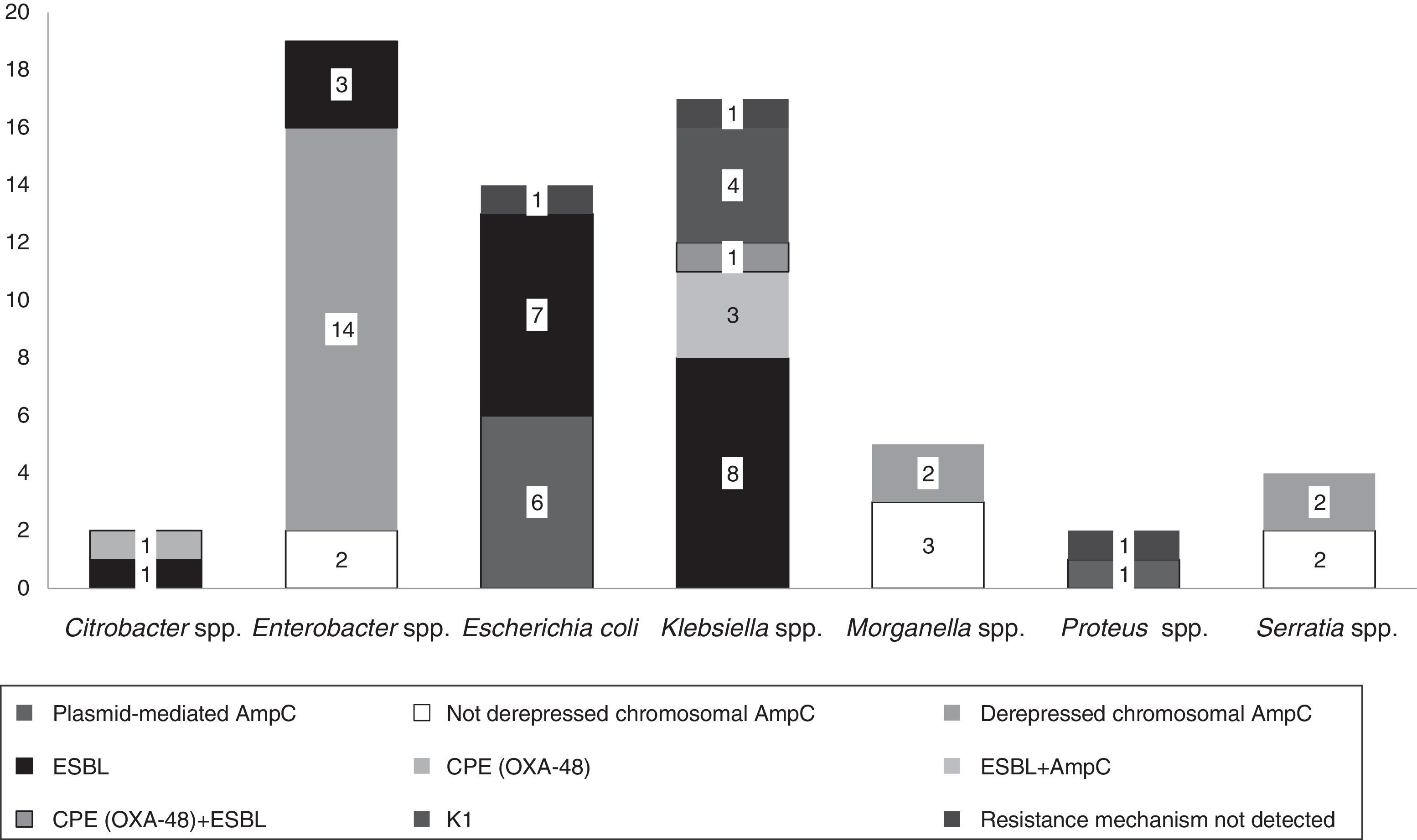
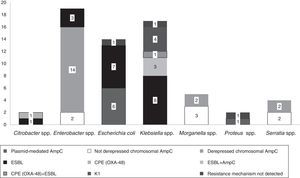
The isolates belonged to the following species: Enterobacter spp. (n=19, 30%), Klebsiella spp. (n=17, 27%), E. coli (n=14, 22%), Morganella spp. (n=5, 8%), Serratia spp. (n=4, 7%), Citrobacter spp. (n=2, 3%) and Proteus spp. (n=2, 3%).
Fifteen isolates had a phenotypic test compatible with a carbapenemase production (synergy with inhibitors employed and/or temocillin resistance). Among them, ten isolates showed synergy with PBA as possible KPC producers and one additional isolate also showed temocillin resistance, most of them corresponded to AmpC β-lactamases inhibited with PBA (Table 1). Four isolates presented temocillin resistance, being two of them confirmed by PCR as OXA-48 producers; the production of carbapenemase was confirmed at the National Reference Laboratory in Spain (Klebsiella pneumoniae 31KP and Citrobacter koseri 52CB) accounting for 3.17% of the strains in this study.
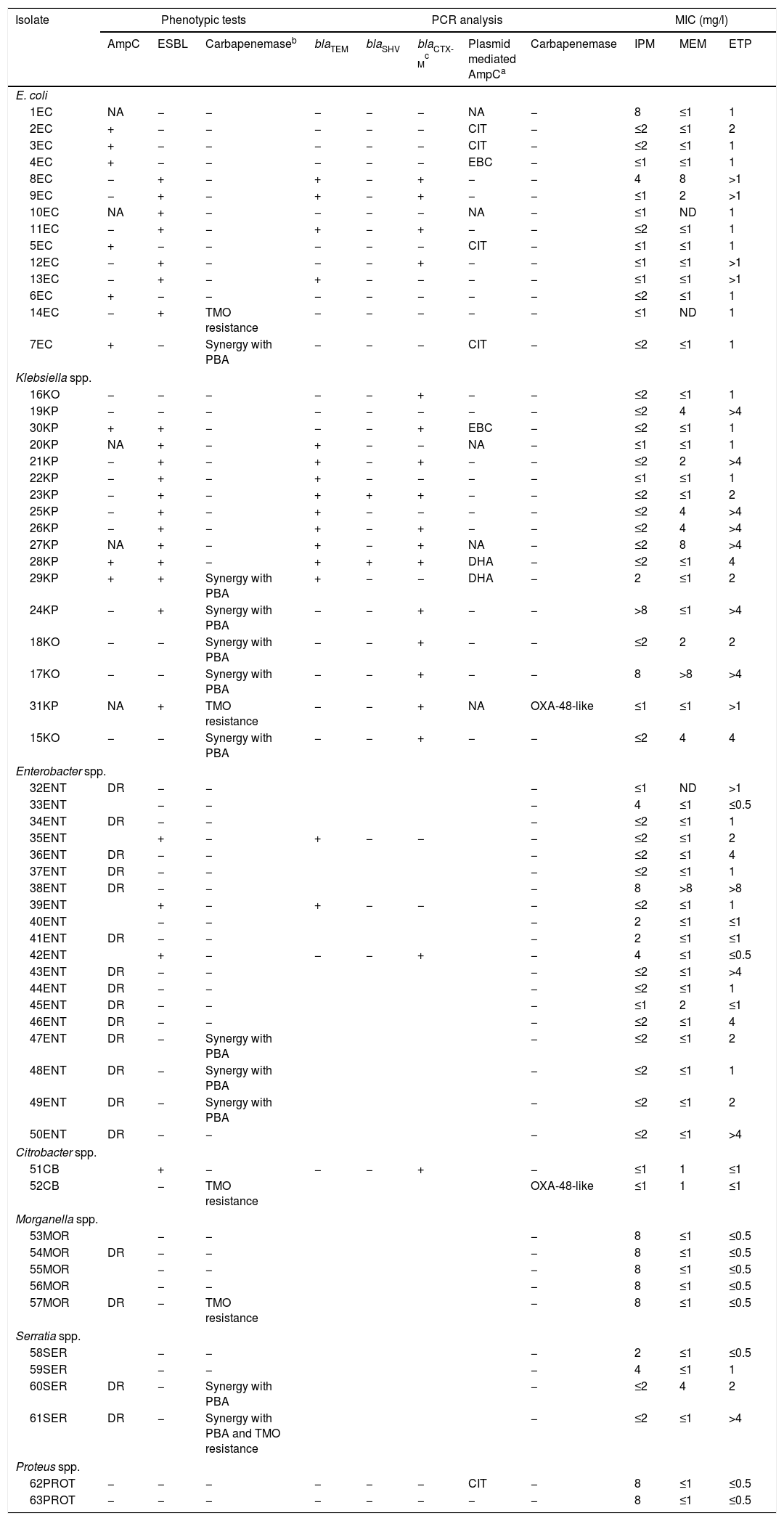
Summary of phenotypic tests, PCR analysis, and carbapenem's MICs against the isolates studied (n=63).
| Isolate | Phenotypic tests | PCR analysis | MIC (mg/l) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AmpC | ESBL | Carbapenemaseb | blaTEM | blaSHV | blaCTX-Mc | Plasmid mediated AmpCa | Carbapenemase | IPM | MEM | ETP | |
| E. coli | |||||||||||
| 1EC | NA | − | − | − | − | − | NA | − | 8 | ≤1 | 1 |
| 2EC | + | − | − | − | − | − | CIT | − | ≤2 | ≤1 | 2 |
| 3EC | + | − | − | − | − | − | CIT | − | ≤2 | ≤1 | 1 |
| 4EC | + | − | − | − | − | − | EBC | − | ≤1 | ≤1 | 1 |
| 8EC | − | + | − | + | − | + | − | − | 4 | 8 | >1 |
| 9EC | − | + | − | + | − | + | − | − | ≤1 | 2 | >1 |
| 10EC | NA | + | − | − | − | − | NA | − | ≤1 | ND | 1 |
| 11EC | − | + | − | + | − | + | − | − | ≤2 | ≤1 | 1 |
| 5EC | + | − | − | − | − | − | CIT | − | ≤1 | ≤1 | 1 |
| 12EC | − | + | − | − | − | + | − | − | ≤1 | ≤1 | >1 |
| 13EC | − | + | − | + | − | − | − | − | ≤1 | ≤1 | >1 |
| 6EC | + | − | − | − | − | − | − | − | ≤2 | ≤1 | 1 |
| 14EC | − | + | TMO resistance | − | − | − | − | − | ≤1 | ND | 1 |
| 7EC | + | − | Synergy with PBA | − | − | − | CIT | − | ≤2 | ≤1 | 1 |
| Klebsiella spp. | |||||||||||
| 16KO | − | − | − | − | − | + | − | − | ≤2 | ≤1 | 1 |
| 19KP | − | − | − | − | − | − | − | − | ≤2 | 4 | >4 |
| 30KP | + | + | − | − | − | + | EBC | − | ≤2 | ≤1 | 1 |
| 20KP | NA | + | − | + | − | − | NA | − | ≤1 | ≤1 | 1 |
| 21KP | − | + | − | + | − | + | − | − | ≤2 | 2 | >4 |
| 22KP | − | + | − | + | − | − | − | − | ≤1 | ≤1 | 1 |
| 23KP | − | + | − | + | + | + | − | − | ≤2 | ≤1 | 2 |
| 25KP | − | + | − | + | − | − | − | − | ≤2 | 4 | >4 |
| 26KP | − | + | − | + | − | + | − | − | ≤2 | 4 | >4 |
| 27KP | NA | + | − | + | − | + | NA | − | ≤2 | 8 | >4 |
| 28KP | + | + | − | + | + | + | DHA | − | ≤2 | ≤1 | 4 |
| 29KP | + | + | Synergy with PBA | + | − | − | DHA | − | 2 | ≤1 | 2 |
| 24KP | − | + | Synergy with PBA | − | − | + | − | − | >8 | ≤1 | >4 |
| 18KO | − | − | Synergy with PBA | − | − | + | − | − | ≤2 | 2 | 2 |
| 17KO | − | − | Synergy with PBA | − | − | + | − | − | 8 | >8 | >4 |
| 31KP | NA | + | TMO resistance | − | − | + | NA | OXA-48-like | ≤1 | ≤1 | >1 |
| 15KO | − | − | Synergy with PBA | − | − | + | − | − | ≤2 | 4 | 4 |
| Enterobacter spp. | |||||||||||
| 32ENT | DR | − | − | − | ≤1 | ND | >1 | ||||
| 33ENT | − | − | − | 4 | ≤1 | ≤0.5 | |||||
| 34ENT | DR | − | − | − | ≤2 | ≤1 | 1 | ||||
| 35ENT | + | − | + | − | − | − | ≤2 | ≤1 | 2 | ||
| 36ENT | DR | − | − | − | ≤2 | ≤1 | 4 | ||||
| 37ENT | DR | − | − | − | ≤2 | ≤1 | 1 | ||||
| 38ENT | DR | − | − | − | 8 | >8 | >8 | ||||
| 39ENT | + | − | + | − | − | − | ≤2 | ≤1 | 1 | ||
| 40ENT | − | − | − | 2 | ≤1 | ≤1 | |||||
| 41ENT | DR | − | − | − | 2 | ≤1 | ≤1 | ||||
| 42ENT | + | − | − | − | + | − | 4 | ≤1 | ≤0.5 | ||
| 43ENT | DR | − | − | − | ≤2 | ≤1 | >4 | ||||
| 44ENT | DR | − | − | − | ≤2 | ≤1 | 1 | ||||
| 45ENT | DR | − | − | − | ≤1 | 2 | ≤1 | ||||
| 46ENT | DR | − | − | − | ≤2 | ≤1 | 4 | ||||
| 47ENT | DR | − | Synergy with PBA | − | ≤2 | ≤1 | 2 | ||||
| 48ENT | DR | − | Synergy with PBA | − | ≤2 | ≤1 | 1 | ||||
| 49ENT | DR | − | Synergy with PBA | − | ≤2 | ≤1 | 2 | ||||
| 50ENT | DR | − | − | − | ≤2 | ≤1 | >4 | ||||
| Citrobacter spp. | |||||||||||
| 51CB | + | − | − | − | + | − | ≤1 | 1 | ≤1 | ||
| 52CB | − | TMO resistance | OXA-48-like | ≤1 | 1 | ≤1 | |||||
| Morganella spp. | |||||||||||
| 53MOR | − | − | − | 8 | ≤1 | ≤0.5 | |||||
| 54MOR | DR | − | − | − | 8 | ≤1 | ≤0.5 | ||||
| 55MOR | − | − | − | 8 | ≤1 | ≤0.5 | |||||
| 56MOR | − | − | − | 8 | ≤1 | ≤0.5 | |||||
| 57MOR | DR | − | TMO resistance | − | 8 | ≤1 | ≤0.5 | ||||
| Serratia spp. | |||||||||||
| 58SER | − | − | − | 2 | ≤1 | ≤0.5 | |||||
| 59SER | − | − | − | 4 | ≤1 | 1 | |||||
| 60SER | DR | − | Synergy with PBA | − | ≤2 | 4 | 2 | ||||
| 61SER | DR | − | Synergy with PBA and TMO resistance | − | ≤2 | ≤1 | >4 | ||||
| Proteus spp. | |||||||||||
| 62PROT | − | − | − | − | − | − | CIT | − | 8 | ≤1 | ≤0.5 |
| 63PROT | − | − | − | − | − | − | − | − | 8 | ≤1 | ≤0.5 |
ETP: ertapenem; IPM: imipenem; MEM: meropenem; TMO: temocillin; −: absence; +: presence; DR: derepressed AmpC; ND: not determined; NA: not available; PBA: phenil-boronic acid.
EC: Escherichia coli; KO: Klebsiella oxytoca; KP: Klebsiella pneumoniae; ENT: Enterobacter spp.; CB: Citrobacter spp.; MOR: Morganella spp.; SER: Serratias spp.; PROT: Proteus spp.
Phenotypic ESBL detection was positive in 27 isolates; beta-lactamase-encoding genes were detected in 25 of them (39.6%), being blaTEM detected in fifteen isolates (60%, 15/25), blaCTX-M in fourteen (56%, 14/25) and blaSHV in two (8%, 2/25). Eight isolates combined more than one resistance beta-lactamase gene (Table 1).
In 14.2% (n=9) of the isolates, plasmid-mediated AmpC beta-lactamases were detected, most of them (n=5) amplified with CIT primers (families LAT-1 to LAT-4, CMY-2 to CMY-7 and BIL-1). Derepressed AmpC β-lactamase, according to reduced susceptibility to third-generation cephalosporins (cefotaxime, ceftriaxone and ceftazidime) and monobactams (aztreonam)12 was present in 18 isolates (28%). No beta-lactamase genes were detected by PCR in three isolates (K. pneumoniae 19KP, E. coli 1EC and Proteus vulgaris 63PROT). Isolate 19KP showed a cefotaxime MIC=2mg/l, ceftazidime MIC=16mg/l, cefepime MIC >16mg/l, aztreonam MIC=8mg/l, meropenem MIC=4mg/l and ertapenem MIC >4mg/l and no AmpC or ESBL confirmation was observed by phenotypic and genotypic methods. Isolate 1EC was susceptible to cephalosporins (no ESBL or AmpC-type) but showed high carbapenem MICs (imipenem MIC=8mg/l, ertapenem MIC=1mg/l), and isolate 63PROT showed resistance to cefotaxime (MIC >32mg/l), compatible with CumA betalactamase, and to imipenem (MIC=8mg/l) (Fig. 1).
According to EUCAST breakpoints, among our isolates analyzed, 20.6% (13/63) were non-susceptible to imipenem, 14.2% (9/63) to meropenem, and 84.1% (53/63) to ertapenem. Two isolates (K. oxytoca 17KO with hyperproduction of the chromosomally encoded K1 β-lactamase and Enterobacter spp., 38ENT showing derepressed chromosomal AmpC) were resistant to all three antibiotics assayed (Table 1).
DiscussionThe emergence of CPE as well as the resistance to carbapenems is a cause of public health concern causing community and nosocomially acquired-infections. This study contributes to clarify the mechanisms involved in reduced susceptibility to carbapenems. Despite several outbreaks described around our country, CPE have not been detected in our hospital until 2014 where two cases of OXA-48-like CPE were confirmed.
Our results show that most of our carbapenem non-susceptible isolates (82.5%) were ESBL or AmpC producers, probably combined with decreased permeability of the outer membrane (porin loss or mutation of the porin-coding sequence) and/or overexpression of efflux pumps. The contribution to carbapenem resistance by loss of porins has been widely described.13–15 Unfortunately, in our laboratory it is not possible to analyze this mechanism of resistance and this is one limitation of our work. Further studies will be needed to analyze this mechanism. In contrast, the role of efflux pumps as contributors to resistance to carbapenems in Enterobacteriaceae is infrequent.16
Most of the AmpC producers showed PBA synergy in the carbapenemase phenotypic detection test, a limitation that can be solved by using Mueller Hinton cloxacillin agar.6
All isolates studied expressed a β-lactam resistance mechanism that could be detected or explained, except for three isolates in which no PCR amplicons for carbapenemases or beta-lactamases were obtained. These strains could have another resistance mechanisms responsible for carbapenem resistance, like an expression of different ESBL encoding genes, or a blocker of the porin channel as it has been reported.4
Previous studies4,16 reported an ESBL prevalence close to 50% among isolates with decreased susceptibility to carbapenems; in our case ESBL were phenotypically detected in 42.8% of the isolates and in 39.6% by PCR amplification of the beta-lactamase-encoding genes. It was not the aim of this study to analyze the families to which these ESBL and/or plasmid-mediated AmpC isolates with a negative PCR belonged; this strategy would require complete gene sequencing analysis. Nevertheless, taking into account the results obtained, 52% of the phenotypic ESBL isolates belonged to CTX-M family, which has been described as the most prevalent worldwide,17 and most of AmpC amplification products observed could be according to the prevalent family in our area CMY-2.17,18
In our study, nineteen chromosomal AmpC isolates showed resistance to cefepime. This resistance could be due to AmpC hyperproduction associated with porin loss or to AmpC with hydrolytic activity against cefepime, as previously reported.19 The fact that CPE may show a low level of resistance or even in vitro susceptibility to carbapenems (not detectable with commercial antimicrobial susceptibility testing panels, as in our study) suggests that active surveillance of these enzymes is critical in order to avoid misdiagnosis and/or underestimation of its prevalence.
The phenotypic detection of carbapenemases (mainly OXA-48-like carbapenemases) remains challenging, due to the low level of resistance sometimes presented, and the possibility of other resistance mechanisms like ESBL or AmpC combined with porine loss. Nevertheless, given the existing situation in surrounding areas it is necessary to strengthen the surveillance, especially in patients with risk factors.
Conflicts of interestThe authors declare no conflicts of interest.
We thank Marina Oviaño for advice and support given.