Most microbiology laboratories use different techniques for the diagnosis of gastrointestinal infections. Some of which require at least 72h to obtain final results.
Material and methodsThe gastrointestinal panel Luminex (xTAG-GPP, Luminex Molecular Diagnostics, Toronto, Canada) is a qualitative multiplex fast and sensitive assay able to detect and to identify the 15 most common pathogens causing gastrointestinal infection simultaneously. We evaluated this multiplex panel comparing it with conventional methods used in our laboratory.
ResultsWe analysed 225 samples of faeces. Through the conventional methods were positive 74 samples (32.9%). Through the Luminex method were positive 137 samples (60.9%).
ConclusionsThe use of the xTAG® GPP system in Clinical Microbiology can improve the diagnosis of gastrointestinal infectious because it provides results in less than 8h. Some pathogens should be applied with caution and should be interpreted based on the patient's clinical data.
La mayoría de laboratorios de microbiología utilizan diferentes técnicas para el diagnóstico de las infecciones gastrointestinales. Algunas requieren hasta 72 h para obtener resultados definitivos.
Material y métodosEl panel gastrointestinal de Luminex (xTAG-GPP, Luminex Molecular Diagnostics, Toronto, Canadá) se trata de un ensayo cualitativo multiplex rápido y sensible capaz de detectar e identificar simultáneamente los 15 patógenos más frecuentes causantes de gastroenteritis. Nuestro objetivo ha sido evaluar este panel multiplex comparándolo con los métodos habituales empleados en nuestro laboratorio.
ResultadosSe analizaron 225 muestras de heces. A través de los métodos convencionales fueron 74 las muestras positivas (32,9%). A través de Luminex fueron 137 las muestras positivas (60,9%).
ConclusiónEl uso del panel gastrointestinal de Luminex puede mejorar el diagnóstico de las infecciones gastrointestinales principalmente porque proporciona resultados en menos de 8h. Determinados microorganismos deben interpretarse con precaución y basándose en datos clínicos y epidemiológicos del paciente.
Gastrointestinal infections cause high morbidity in our area. A wide spectrum of microorganisms can be responsible and present the same clinical symptoms. Therefore, the identification of the aetiological agent is important for diagnosis and treatment. The majority of microbiology laboratories use different techniques for diagnosis and some, such as culturing, need up to 72h to obtain definitive results. As an alternative, certain laboratories have molecular methods that are used in specific situations, but these detect a limited number of microorganisms in a single analysis.
The xTAG-GPP Gastrointestinal Pathogen Panel is a qualitative multiplex PCR assay which is able to simultaneously detect and identify, in less than 8h, the 15 most common pathogens that cause gastroenteritis. The included pathogens are: (a) 3 viruses: Adenovirus 40/41, Rotavirus A, Norovirus GI/GII; (b) 9 bacteria: Salmonella spp., Shigella spp., Vibrio cholerae, Yersinia enterocolitica, Campylobacter jejuni, Escherichia coli O 157 and toxins such as Clostridium difficile toxins A/B, enterotoxigenic E. coli (ETEC): heat-labile enterotoxin (LT)/heat-stable enterotoxin (ST), E. coli O 157, Shiga toxin-producing E. coli (STEC stx1/stx2) and (c) 3 parasites: Cryptosporidium spp., Entamoeba histolytica and Giardia lamblia.
Our main objective was to compare the results obtained via conventional diagnostic methods used in our laboratory with those obtained via the Luminex molecular panel.
Material and methodsWe analysed 225 fresh faecal samples submitted in sterile screw-top containers, from patients with acute diarrhoea and of any age. The samples were sent to the Microbiology Department of Consorcio Hospital General Universitario de Valencia from July to November 2013, following the appropriate rules for shipping and storage.
Initially the samples were processed routinely. The culture was used to detect Salmonella spp., Shigella spp., Y. enterocolitica, Campylobacter spp., Aeromonas spp. and Vibrio spp. For that purpose, low selective media, such as MacConkey agar and blood agar, culture media with mid selectivity such as Hektoen agar, and enriched fluid media, such as selenite broth, were used. They were all incubated for 24h at 37°C, except the selenite broth, which was incubated for 48h. To isolate Campylobacter spp. a selective medium was used and it was incubated for 48h in microaerophilic conditions at a temperature of 42°C. For identification, the suspected colonies were analysed with the MALDI-TOF system and for the susceptibility test, the MicroScan (Siemens) NC69 panel was used. Toxigenic C. difficile was detected via enzyme immunoassay (C. difficile Quik Chek Complete, Alere). This test detects the antigen glutamate dehydrogenase and toxins A/B directly from faecal samples. The positive toxin A/B results were confirmed with PCR, as indicated by the protocols. A rapid faecal antigen detection test (immunochromatography, Rota-Adeno Letitest) was used to detect Adenovirus and Rotavirus. Norovirus is not routinely studied in our hospital. The presence of parasites is performed using microscopy techniques.
Immediately after their usual processing, the faeces were stored at −80°C until later analysis using the Luminex system. This technique includes various steps: sample pre-treatment, extraction of nucleic acid, multiplex RT-PCR, hybridisation reaction with microspheres and a detection and interpretation phase using the Luminex instrument (Luminex Molecular Diagnostics, Toronto, Canada) via the TDAS GPP 1.11 software program.
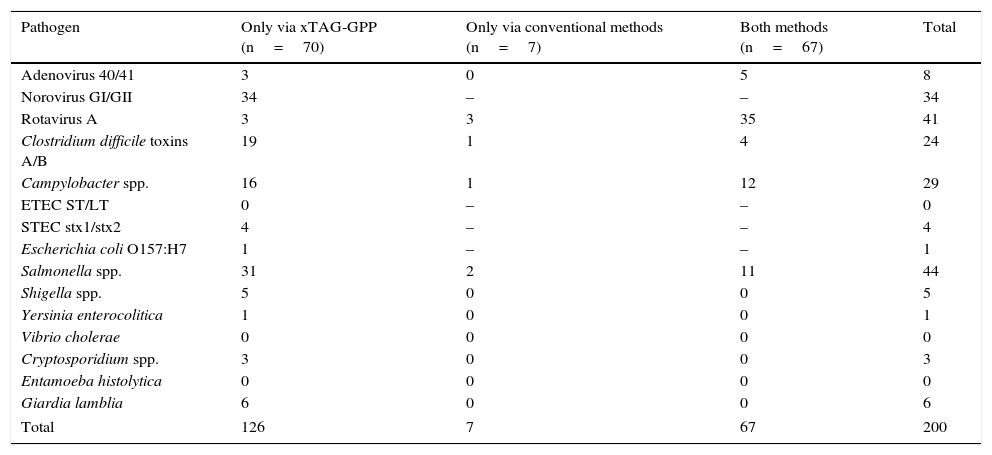
Results225 faecal samples were analysed, and in 144 (64%) one or more pathogens were detected (Table 1). The positive samples for which results were consistent with both methods were 67/225 (29.7%). Rotavirus A was the most common pathogen.
Pathogens detected by method used (n=144).
| Pathogen | Only via xTAG-GPP (n=70) | Only via conventional methods (n=7) | Both methods (n=67) | Total |
|---|---|---|---|---|
| Adenovirus 40/41 | 3 | 0 | 5 | 8 |
| Norovirus GI/GII | 34 | – | – | 34 |
| Rotavirus A | 3 | 3 | 35 | 41 |
| Clostridium difficile toxins A/B | 19 | 1 | 4 | 24 |
| Campylobacter spp. | 16 | 1 | 12 | 29 |
| ETEC ST/LT | 0 | – | – | 0 |
| STEC stx1/stx2 | 4 | – | – | 4 |
| Escherichia coli O157:H7 | 1 | – | – | 1 |
| Salmonella spp. | 31 | 2 | 11 | 44 |
| Shigella spp. | 5 | 0 | 0 | 5 |
| Yersinia enterocolitica | 1 | 0 | 0 | 1 |
| Vibrio cholerae | 0 | 0 | 0 | 0 |
| Cryptosporidium spp. | 3 | 0 | 0 | 3 |
| Entamoeba histolytica | 0 | 0 | 0 | 0 |
| Giardia lamblia | 6 | 0 | 0 | 6 |
| Total | 126 | 7 | 67 | 200 |
Using the Luminex method, 137/225 (60.9%) of the samples were positive, but only 70/225 (31.1%) were positive solely via this method. The “extra” pathogens detected were: Norovirus (n=34), Adenovirus and Rotavirus (n=3), Salmonella spp. (n=31), followed by toxigenic C. difficile (n=19) and Campylobacter spp. (n=16). Shigella spp. was present in 5 samples and Yersinia spp., in one sample. Four samples were positive for STEC and one for E. coli serotype O 157. V. cholerae, ETEC and E. histolytica were not found in any sample. Cryptosporidium spp. and G. lamblia were present in 3 and 6 samples, respectively.
Using the conventional methods, 74/225 (32.9%) were positive, but only 7 samples were positive solely via this method (3.1%). The involved microorganisms were: Rotavirus A (n=3), Salmonella spp. (n=2), Campylobacter spp. and toxigenic C. difficile (n=1).
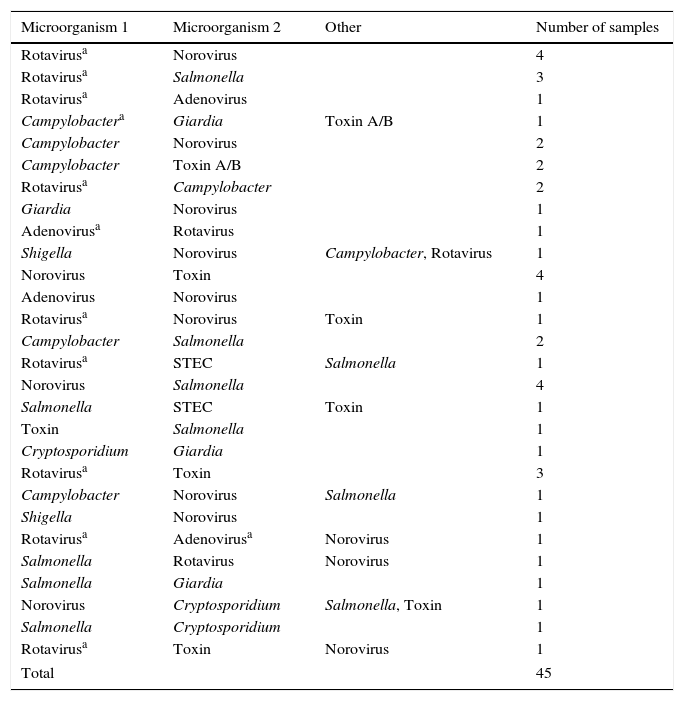
In 45 samples (20%), more than one pathogen was detected via Luminex. The microorganisms and combinations found are listed in Table 2.
Combinations detected (n=45).
| Microorganism 1 | Microorganism 2 | Other | Number of samples |
|---|---|---|---|
| Rotavirusa | Norovirus | 4 | |
| Rotavirusa | Salmonella | 3 | |
| Rotavirusa | Adenovirus | 1 | |
| Campylobactera | Giardia | Toxin A/B | 1 |
| Campylobacter | Norovirus | 2 | |
| Campylobacter | Toxin A/B | 2 | |
| Rotavirusa | Campylobacter | 2 | |
| Giardia | Norovirus | 1 | |
| Adenovirusa | Rotavirus | 1 | |
| Shigella | Norovirus | Campylobacter, Rotavirus | 1 |
| Norovirus | Toxin | 4 | |
| Adenovirus | Norovirus | 1 | |
| Rotavirusa | Norovirus | Toxin | 1 |
| Campylobacter | Salmonella | 2 | |
| Rotavirusa | STEC | Salmonella | 1 |
| Norovirus | Salmonella | 4 | |
| Salmonella | STEC | Toxin | 1 |
| Toxin | Salmonella | 1 | |
| Cryptosporidium | Giardia | 1 | |
| Rotavirusa | Toxin | 3 | |
| Campylobacter | Norovirus | Salmonella | 1 |
| Shigella | Norovirus | 1 | |
| Rotavirusa | Adenovirusa | Norovirus | 1 |
| Salmonella | Rotavirus | Norovirus | 1 |
| Salmonella | Giardia | 1 | |
| Norovirus | Cryptosporidium | Salmonella, Toxin | 1 |
| Salmonella | Cryptosporidium | 1 | |
| Rotavirusa | Toxin | Norovirus | 1 |
| Total | 45 | ||
The Luminex panel presents various advantages over conventional methods. The most highlighted is the simultaneous detection of multiple pathogens and the decrease in diagnostic time to 6–8h.1–6 Also, it allows for detection of microorganisms for which we do not have routine diagnostic methods, and of mixed infections.
In this study, a total of 126 “extra” pathogens were detected. Salmonella spp. and Norovirus were the predominant pathogens, followed by C. difficile toxin and Campylobacter spp. In the case of Salmonella, in our area a precise diagnosis is very important. Its high and unexpected detection via the Luminex panel has also been highlighted in other articles.1–3,7 The same situation is described for E. histolytica in regions where this parasite is highly prevalent.1,7,8 In our study, however, it was not detected in any samples.
In the detection of C. difficile, 69.5% (16/23) belonged to faecal samples from children under the age of 5. That is, it was detected in a group of the population where the rapid test had not been requested by the clinician. In the paediatric population this is still a topic of debate due to the high possibility of asymptomatic carriers. Even so, as Khare et al.9 indicate, the high detection of C. difficile toxin in children represents an important area of study.
Norovirus was also detected in a large number of samples (n=34). Many laboratories do not routinely test for this virus due to the low sensitivity of EIA methods. Therefore, it is suspected to be the cause of a large number of undiagnosed cases. In this study, antigen detection was not conducted and, therefore, we lack the data to be able to compare them. Nevertheless, unexpected results, both for Norovirus and for Campylobacter have been reported by other authors.3,4
In our study, the detection of Rotavirus via Luminex, when comparing them with the immunological antigen detection methods, we observed that 85% (35/41) of the cases had consistent results.
Less common pathogens, such as Shigella spp., Y. enterocolitica, STEC, Cryptosporidium spp. and G. lamblia, were detected in different samples by Luminex, unlike the usual methods. We do not have results with a second molecular method. Therefore, we cannot rule out the possibility of false positives. In publications by other authors, such as Beckmann et al.,4 half of the cases in which G. lamblia was detected were not confirmed. In the case of Shigella, 5 patients were involved, and we did check them with a simple PCR. Two cases were confirmed, for which we had access to the medical records, and both had recently travelled to India.
In a minority of samples (n=7), the discrepancies were due to the fact that they were positive using routine methods, but negative using Luminex. One possible cause would be degradation of the nucleic acids despite optimal storage of the sample at −80°C.
Mixed infections are not common. In this study, more than one pathogen was detected in 45 samples (20%). The combinations change from one study to another,2,4,9–12 probably because the geographical area and population type vary.
The high sensitivity of the technique and the wide detection of microorganisms in a single analysis provide a lot of information, which is sometimes difficult to interpret. Some studies have indicated the need to quantify enteropathogens, especially in mixed infections, and to differentiate between pathogenic colonisation or active replication in order to determine the implications of each one,12 since it is known that certain microorganisms can be present in asymptomatic patients or a long period of time after the resolution of the infection.
With regard to other molecular panels marketed and approved for use in clinical practice,13 Luminex has a high yield because it has the capacity to analyse up to 24 samples in a single panel, in addition to being one of the panels that detects the greatest number of pathogens per analysis. But it is a laborious and complex technique that requires qualified staff experienced in molecular biology techniques. Furthermore, there is a high risk of contaminations, since it is an open system. And daily maintenance and meticulous cleaning is essential, both of the instrument and the work site. All of this complicates its implementation as a routine method in most microbiology laboratories, in addition to the fact that it would not replace culturing, since in the case of positive samples, confirmation or having the strain available for susceptibility studies would be necessary. Finally, its higher cost compared to other methods should be mentioned. To obtain an optimal cost/benefit ratio, a high number of samples must be analysed, and this is not feasible in many laboratories. However, an in-depth study would be needed for this purpose.
In conclusion, the multiplex Luminex Gastrointestinal Pathogen Panel provides major advantages in the diagnosis of acute gastroenteritis since it is fast and identifies the pathogens causing 80% of infectious gastroenteritis in a single analysis. Positive results for particular pathogens should be interpreted with caution and within the patient's clinical context. We recommend its use mainly in outbreaks, epidemiological studies, in cases of travellers’ diarrhoea and in seriously ill patients where there is a high suspicion of infectious disease.
Conflicts of interestThe authors declare no conflicts of interest.
The authors thank Luminex Molecular Diagnostic for having provided the instrument and reagents used in this study.
Please cite this article as: Casañ C, Ocete MD, Medina R, Gimeno C. Evaluación del panel gastrointestinal xTAG®-GPP de Luminex en el diagnóstico de las gastroenteritis agudas. Enferm Infecc Microbiol Clin. 2017;35:574–577.