The aim was to evaluate a rapid method which would combine identification and susceptibility testing directly from positive blood cultures for Gram-negative bacilli of the Enterobacterales.
Material and methodsGram-negative rods from blood cultures were directly identified by MALDI-TOF. Samples with Enterobacterales were selected for direct antimicrobial susceptibility testing by Vitek 2. The results were compared to those obtained with our laboratory's standard method.
ResultsMALDI-TOF directly from blood cultures identified correctly 83% of the samples. Enterobacterales (n=68) were identified at gender and species level in 85% of blood cultures with a score >1.7. In general, MICs were obtained after 7h. MICs of amoxicillin-clavulanate, amikacin and ciprofloxacin showed in almost 50% of the cases after 5h.
ConclusionsA simple procedure with low cost and reduced working time makes it possible to integrate both identification and susceptibility testing directly from blood cultures. Thus, this protocol could offer advantages when it comes to selection and cost of treatment and patients’ clinical outcomes.
Evaluar un método rápido de identificación y estudio de sensibilidad directamente desde hemocultivos positivos para bacilos gramnegativos del orden Enterobacterales.
Material y métodosLos hemocultivos con bacilos gramnegativos fueron utilizados para identificación mediante MALDI-TOF, seleccionándose aquellos con Enterobacterales para estudio de sensibilidad con Vitek 2. Los resultados fueron comparados con el método estándar del laboratorio.
ResultadosMALDI-TOF identificó correctamente el 83% de las muestras obtenidas directamente de hemocultivos. En caso de Enterobacterales (n=68), el 85% se identificaron a nivel de género y especie con un score>1,7. El tiempo necesario para la obtención de CMIs fue de 7 horas, reduciéndose a 5 en amoxicilina-clavulánico, amicacina y ciprofloxacino, casi en el 50% de los casos.
ConclusionesUn protocolo simplificado, con bajo coste y reducido tiempo de trabajo, combinando identificación y estudio de sensibilidad directamente desde hemocultivos, podría proporcionar beneficios en la elección y coste del tratamiento, y mejora clínica del paciente.
Bloodstream infections are associated with high rates of morbidity and mortality, with 19 million cases every year.1,2 Appropriate therapy reduces mortality, treatment costs and improves clinical outcomes.3,4 Classical methods for identification (ID) and antimicrobial susceptibility testing (AST) require 3–4 days from the extraction of the sample to obtain results. Several methods have been evaluated in order to shorten this time.2,5,6 The aim of this study is to evaluate a method of ID and AST directly from positive blood cultures (BCs), with a short hands-on time and a little volume of sample.
Material and methodsA total of 103 positive BCs showing gram-negative rods (GNR) were analysed for rapid ID by MALDI-TOF (Bruker Daltonics). Microorganisms belonging to Enterobacterales were processed for direct AST by Vitek 2 (BioMerieux). One BC per patient was included. BCs with polymicrobial growth were excluded. Results obtained directly from positive BCs were compared with the methods used in our laboratory routine.
BC bottles were incubated at 35°C for 5 days in an automated system (Becton Dickinson). Positivity of BCs was confirmed by Gram staining and were then subcultured on different agar solid media. At the same time, an aliquot of the BC was used for direct ID and AST. After 24h of incubation, isolated colonies on solid media were processed for ID by MALDI-TOF, and for ID and AST by Microscan WalkAway (Beckman Coulter).
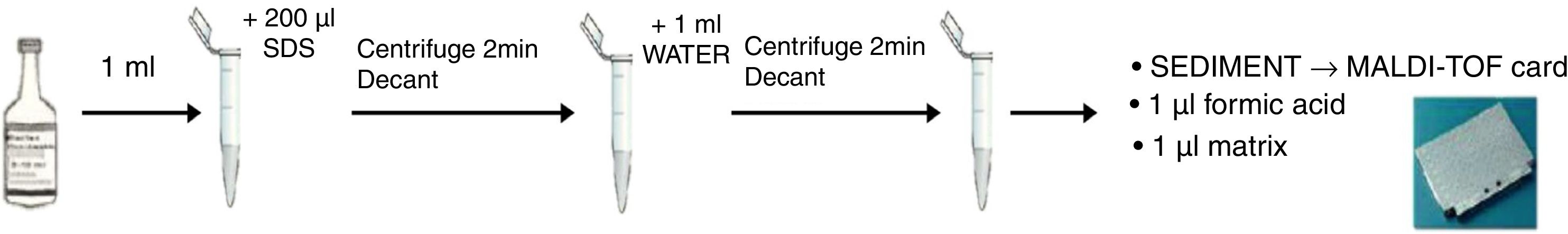
Direct ID by MALDI-TOF was performed following the protocol described by Hoyos-Mallecot et al. 7 with some modifications (Fig. 1): 1ml of blood culture was transferred to an Eppendorf tube with 200μl of 5% sodium dodecyl sulphate, inverted and centrifuged for 2min at 16,000rpm. The supernatant was removed, the sediment was re-suspended in 1ml of sterile distilled water, again centrifuged, and the supernatant removed. For a quick ID, on-plate formic acid (FA) extraction was used. The pellet was transferred directly onto the target plate and after drying, 1μl of 100% FA was added to each spot. It was dried and then 1μl of α-cyano-4-hydroxy-cinnamic-acid matrix was added.
Spectra were analysed using Biotyper 3.0 software® (Bruker Daltonics). As quality control, the solution Bruker's bacterial test standard was used. Results were expressed as log (score) index. Breakpoints for the interpretation of results were assigned as previously published.8–10 Scores >=1.7 were considered of high-confidence (species level), scores 1.4–1.699 were considered of intermediate confidence (only at genus level), and scores <1.4 were considered unacceptable.
Positive BCs with an ID belonging to Enterobacterales were selected for AST by Vitek 2. Other GNR were not included, as well as samples with score <1.4. Using the same pellet, a bacterial suspension 1–1.5 McFarland standard using sodium chloride (0.45%) was used for AST, following BioMerieux recommendations. AST-N243 cards and the bacterial suspension was introduced in the Vitek 2. Antimicrobial agents tested were ampicillin, amoxicillin-clavulanate, piperacillin-tazobactam, cefuroxime, cefoxitin, cefotaxime, ceftazidime, cefepime, ertapenem, imipenem, amikacin, gentamicin, nalidixic acid, ciprofloxacin and trimethoprim-sulfamethoxazole. Results obtained were collected after 5, 6 and 7h of incubation. The final report was obtained after 10h. Escherichia coli ATCC 25922 and Klebsiella pneumoniae ATCC 1705 were used as controls. To confirm that the bacterial suspension fitted the inoculum required, an agar-blood culture colony count was carried out. Minimal inhibitory concentrations (MIC) obtained using both methods were classified following the recommendations of the Clinical Laboratory Standards Institute.11 ID discordant results were confirmed by MALDI-TOF performed from colonies.
Discrepancies in the interpretation of the susceptibility were analysed and classified in three groups: very major error (susceptible by Vitek 2 but resistant according to the reference method), major error (resistant by Vitek 2 but susceptible by the reference method), and minor error if resistant or susceptible by Vitek 2 and intermediate according to the reference method or vice versa. Errors were verified with gradient diffusion strips (Liofilchem®).
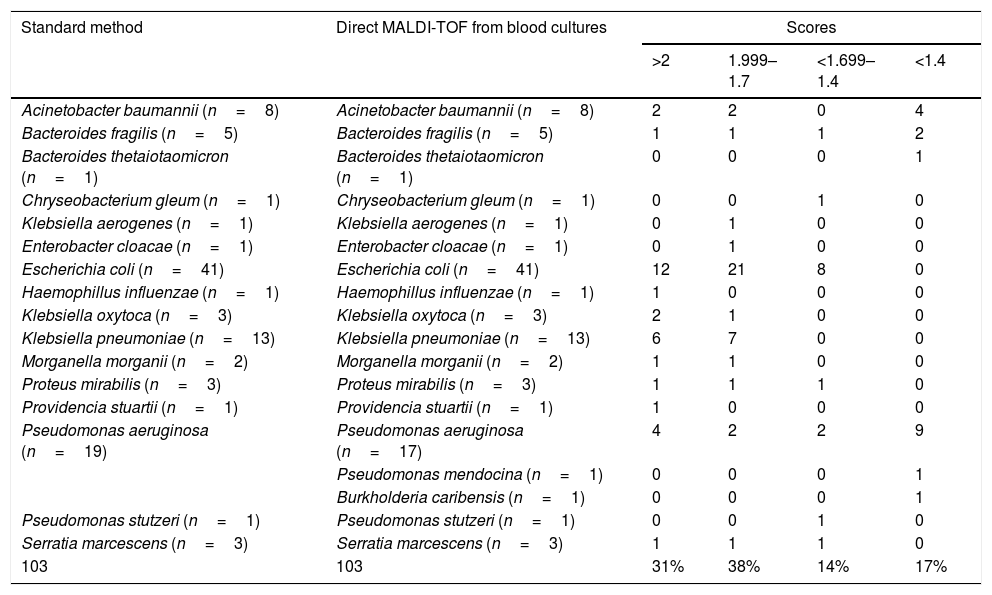
ResultsAll 103 isolates included were GNR, as confirmed by Gram staining. Table 1 shows results obtained by MALDI-TOF directly from BC and the reference methods; 68 of the isolates were Enterobacterales, 29 were non-lactose fermenting GNR, 6 anaerobic GNR and 1 fastidious GNR. Using our method, 83% of the organisms were correctly identified (score >1.4). All Enterobacterales were correctly identified. Two discrepancies were found in the ID of Pseudomonas aeruginosa. One of them was correctly identified at the genus level. The agreement of this method with the standard procedures was 98%. The susceptibility and the predictive positive value were 85% and 98%, respectively.
Identification of gramnegative rods by MALDI-TOF directly from blood cultures vs. standard method.
| Standard method | Direct MALDI-TOF from blood cultures | Scores | |||
|---|---|---|---|---|---|
| >2 | 1.999–1.7 | <1.699–1.4 | <1.4 | ||
| Acinetobacter baumannii (n=8) | Acinetobacter baumannii (n=8) | 2 | 2 | 0 | 4 |
| Bacteroides fragilis (n=5) | Bacteroides fragilis (n=5) | 1 | 1 | 1 | 2 |
| Bacteroides thetaiotaomicron (n=1) | Bacteroides thetaiotaomicron (n=1) | 0 | 0 | 0 | 1 |
| Chryseobacterium gleum (n=1) | Chryseobacterium gleum (n=1) | 0 | 0 | 1 | 0 |
| Klebsiella aerogenes (n=1) | Klebsiella aerogenes (n=1) | 0 | 1 | 0 | 0 |
| Enterobacter cloacae (n=1) | Enterobacter cloacae (n=1) | 0 | 1 | 0 | 0 |
| Escherichia coli (n=41) | Escherichia coli (n=41) | 12 | 21 | 8 | 0 |
| Haemophillus influenzae (n=1) | Haemophillus influenzae (n=1) | 1 | 0 | 0 | 0 |
| Klebsiella oxytoca (n=3) | Klebsiella oxytoca (n=3) | 2 | 1 | 0 | 0 |
| Klebsiella pneumoniae (n=13) | Klebsiella pneumoniae (n=13) | 6 | 7 | 0 | 0 |
| Morganella morganii (n=2) | Morganella morganii (n=2) | 1 | 1 | 0 | 0 |
| Proteus mirabilis (n=3) | Proteus mirabilis (n=3) | 1 | 1 | 1 | 0 |
| Providencia stuartii (n=1) | Providencia stuartii (n=1) | 1 | 0 | 0 | 0 |
| Pseudomonas aeruginosa (n=19) | Pseudomonas aeruginosa (n=17) | 4 | 2 | 2 | 9 |
| Pseudomonas mendocina (n=1) | 0 | 0 | 0 | 1 | |
| Burkholderia caribensis (n=1) | 0 | 0 | 0 | 1 | |
| Pseudomonas stutzeri (n=1) | Pseudomonas stutzeri (n=1) | 0 | 0 | 1 | 0 |
| Serratia marcescens (n=3) | Serratia marcescens (n=3) | 1 | 1 | 1 | 0 |
| 103 | 103 | 31% | 38% | 14% | 17% |
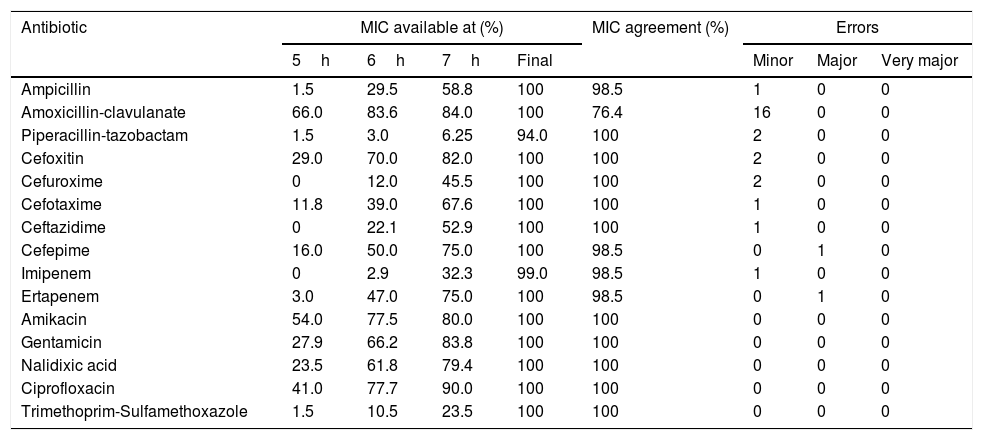
MICs available at each time and for each antimicrobial agent are shown in Table 2, including the final MIC achieved for all antibiotics except piperacillin-tazobactam, which showed results in 94% of the cases. For amoxicillin-clavulanate, amikacin and ciprofloxacin, results were obtained only after 5h of incubation in 50% of the cases. Most MIC results were obtained after 7h of incubation. Trimethoprim-sulfamethoxazole and piperacillin-tazobactam were the only antibiotics whose MIC was available for less than 25% of the isolates after 7h of incubation. Amikacin, ciprofloxacin and trimethoprim-sulfamethoxazole showed 100% agreement with Microscan WalkAway results. Amoxicillin-clavulanate had a 76.4% agreement, whereas ampicillin, cefoxitin, cefuroxime, cefepime and ertapenem showed ≥97% agreement. Twenty-three minor errors were found: 16 of them with amoxicillin-clavulanate. Two major errors were detected for cefepime and ertapenem, and no very major errors were detected at all. The MICs obtained did not change with time and corresponded to the MICs obtained from the colonies using a 0.5 McFarland inoculum.
Percentage of isolates with available MICs at different times of incubation and percentage of MIC agreement and errors between the direct VITEK 2 system and the MicroScan WalkAway (routine standard).
| Antibiotic | MIC available at (%) | MIC agreement (%) | Errors | |||||
|---|---|---|---|---|---|---|---|---|
| 5h | 6h | 7h | Final | Minor | Major | Very major | ||
| Ampicillin | 1.5 | 29.5 | 58.8 | 100 | 98.5 | 1 | 0 | 0 |
| Amoxicillin-clavulanate | 66.0 | 83.6 | 84.0 | 100 | 76.4 | 16 | 0 | 0 |
| Piperacillin-tazobactam | 1.5 | 3.0 | 6.25 | 94.0 | 100 | 2 | 0 | 0 |
| Cefoxitin | 29.0 | 70.0 | 82.0 | 100 | 100 | 2 | 0 | 0 |
| Cefuroxime | 0 | 12.0 | 45.5 | 100 | 100 | 2 | 0 | 0 |
| Cefotaxime | 11.8 | 39.0 | 67.6 | 100 | 100 | 1 | 0 | 0 |
| Ceftazidime | 0 | 22.1 | 52.9 | 100 | 100 | 1 | 0 | 0 |
| Cefepime | 16.0 | 50.0 | 75.0 | 100 | 98.5 | 0 | 1 | 0 |
| Imipenem | 0 | 2.9 | 32.3 | 99.0 | 98.5 | 1 | 0 | 0 |
| Ertapenem | 3.0 | 47.0 | 75.0 | 100 | 98.5 | 0 | 1 | 0 |
| Amikacin | 54.0 | 77.5 | 80.0 | 100 | 100 | 0 | 0 | 0 |
| Gentamicin | 27.9 | 66.2 | 83.8 | 100 | 100 | 0 | 0 | 0 |
| Nalidixic acid | 23.5 | 61.8 | 79.4 | 100 | 100 | 0 | 0 | 0 |
| Ciprofloxacin | 41.0 | 77.7 | 90.0 | 100 | 100 | 0 | 0 | 0 |
| Trimethoprim-Sulfamethoxazole | 1.5 | 10.5 | 23.5 | 100 | 100 | 0 | 0 | 0 |
Our protocol is very promising, with lower costs than other commercial techniques being developed at present. Other advantages are the reduced amount of sample needed and the short hands-on-time to obtain bacterial pellets. ID directly from BCs using MALDI-TOF has proven to be effective and reliable, even though manipulation of the samples requires good knowledge of the procedure. Several methodologies to ID directly from positive BCs have shown optimal results for GNR and problems for grampositive cocci, yeasts and anaerobes. The reasons for the variability of published results are the lack of consensus in the methodology or the breakpoints. Bruker establishes three scores: unreliable identification (<1.7), genus identification (1.7–1.99), or reliable species identification (>2.0). Hoyos-Mallecot et al. demonstrated an improvement of 38% using lower breakpoints.7 Vlek et al. identified correctly 87% of the GNR using lower breakpoints.9 In our case, using lower breakpoints allowed us to correctly identify almost all the BCs processed. Thus, for Enterobacterales it would be necessary to readjust the scores. Other criteria supporting the identification, such as the number of matching identifications, could be used for non-lactose fermenting GNR.
AST with Vitek 2 allows reading MIC results only after 5h of incubation, decreasing the time of response. Besides, MIC does not vary, meaning that a result obtained after a short period of incubation is reliable. This implies an advantage in comparison with a preliminary antibiogram performed from positive BCs, since with the Vitek 2 we can obtain an MIC and detect a mechanism of resistance. This is of paramount importance, as the Vitek 2 gives an alert when resistance to any antimicrobial agent appears, making it easier for the physician to select the proper treatment.
By using MALDI-TOF for direct ID, 100% of Enterobacterales isolates were correctly identified at the genus level, whereas the correct identification for genus and species was achieved in 85% of the cases. Regarding non-lactose fermenting GNR, anaerobes and others, only 50% were correctly identified.
Most MIC agreement errors were minor. In comparative evaluations of susceptibility testing procedures, very major errors should occur only in <1.5% of all tests, and the overall agreement between test and reference method should be superior to 95%.2 In our study we did not find very major errors and complete agreement for all antibiotics was >96%, except for amoxicillin-clavulanate and cefotaxime.
Polymicrobial BCs and low-inoculum size seem to be the two major limitations of our study, together with the identification of non-lactose fermenting GNR and anaerobes, correctly identified only in 50% of the cases. Moreover, this study includes only a few cases of multidrug resistant bacteria, so the conclusions are not applicable for this group of microorganisms.
Additional studies with both non-fermenting GNR and multidrug resistant bacteria are needed, as well as validation studies required to increase the strategy of ID and AST directly from BC in bacteraemia caused by grampositive bacteria and yeasts.
FundingNo funding.
Conflict of interestThe authors affirm that this manuscript is an honest, accurate, and transparent account of the study being reported; that no important aspects of the study have been omitted; and that any discrepancies from the study as planned (and, if relevant, registered) have been explained.
The authors want to thank all the team at the Microbiology Department of Consorcio Hospital General Universitario de Valencia.