Introduction
Hepatitis C virus (HCV) strains are grouped into six main genotypes, namely genotypes 1 to 6, and into several subtypes within most of them 1. Genotype 1a prevails in America and in the North and centre of Europe, whereas genotype 1b prevails in Eastern Europe, the Far East and the Mediterranean. Genotype 3 strains originating from the Indian subcontinent have spread broadly among injection drug users of the Western hemisphere 2. Genotype 4 is thought original from Africa and has shown a significant prevalence in some European countries from the Mediterranean bassin 3. Two main episodes of spread of HCV are thought to have occurred in Europe: the former one involved the spread of genotype 1 strains through blood transfusion, and the second followed the introduction of genotype 3 strains through the use of injection drugs 2.
Genotype 1b has been found prevalent in all studies performed on unselected chronic carriers from Spain, always reaching figures above 50% 4-7. Genotype 3 prevalence ranged from 4 to 17%, rising above 20% among injection drug users 4-8. Prevalence of genotype 4 has been usually found below 10%, but higher figures have been recorded among injection drug users 9-11. These data suggest that the import of genotype 3 strains into Spain would have been accompanied by the introduction and spread of genotype 4. The aim of the present work was to assess changes in the prevalence of HCV genotypes in Spain along the period 1996-2004.
Materials and methods
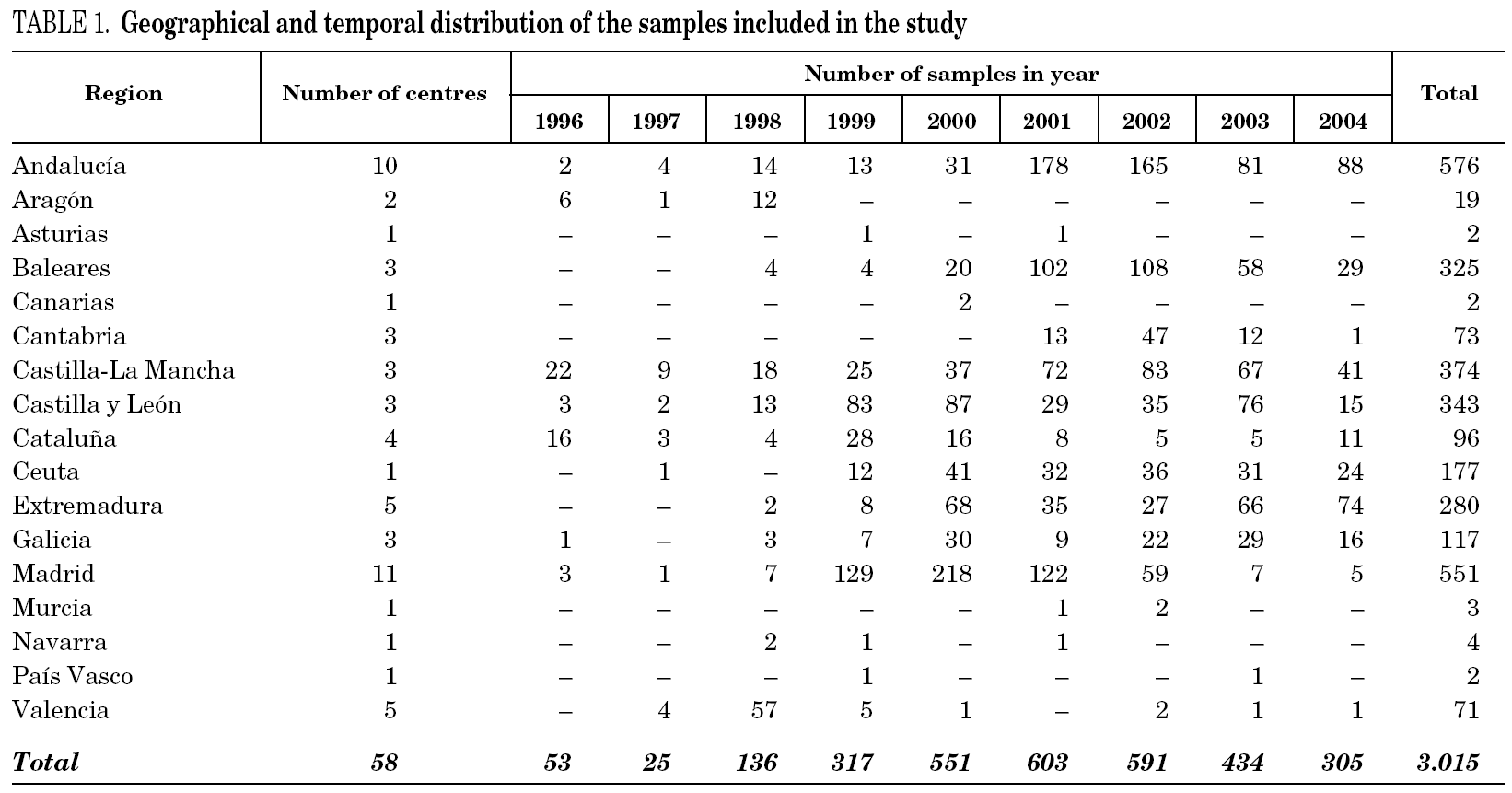
The study was retrospective in nature and included 3161 individual, HCV RNA-positive serum samples from HCV carriers sent to a reference laboratory for genotype identification prior to antiviral treatment. Samples from patients suspected to have been involved in epidemic outbreaks of HCV infection were not included, and no additional selection in regard to age, sex, geographical origin, other risk factors for acquisition of HCV infection or any other parameter was done. After performing genotyping tests, 3015 samples were considered for analysis (see Results).
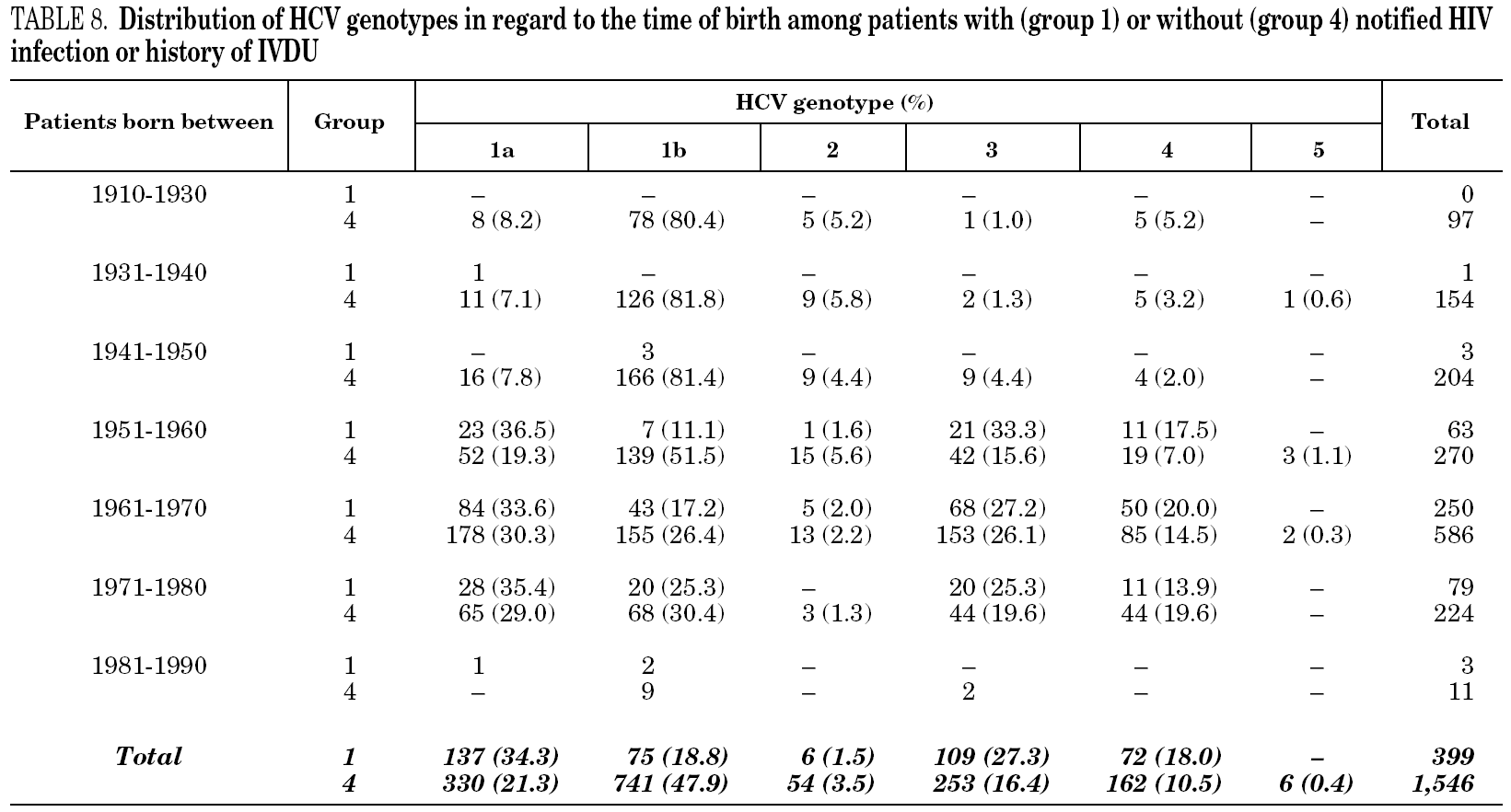
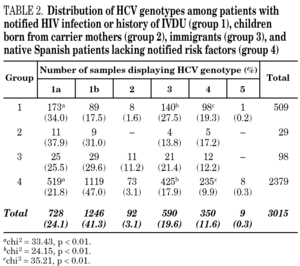
Samples came from 58 health care institutions (21 reference hospitals, 28 regional hospitals, four transfusion centres, four private institutions and one reference laboratory) from 17 different regions of Spain (table 1) and were taken between January, 1996 and December, 2004. Most samples (2743, 90.9%) were from eight regions contributing with more than 100 samples each. Infection by human immunodeficiency virus (HIV) and/or history of intravenous drug use (IVDU) was notified in 509 cases (group 1). Among the remaining, 29 samples were from children born from HCV-infected mothers (group 2); 98 were from immigrants (group 3); and 2379 were from adults native from Spain (group 4). The gender was known from all patients but eight, with a predominance of men (67.8%). Age was known from 2014 patients (66.8%), and ranged from newborn age to 89 years old.
A fragment of the 59 non-coding region of the HCV genome was amplified by the polymerase chain reaction (PCR) 6 from all samples, and the genotype was determined by reverse hybridisation (line probe assay, LiPA), using a commercial test (Versant HCV GenoAmp LiPA, Bayer Healthcare LLC, Tarytown, NY, USA). Prevalence of genotypes 1a, 1b, 2, 3, 4, 5 and 6 was calculated, and results from samples grouped by different criteria were compared by the chi square (chi 2) test. Differences were considered statistically significant for p < 0,01.
Results
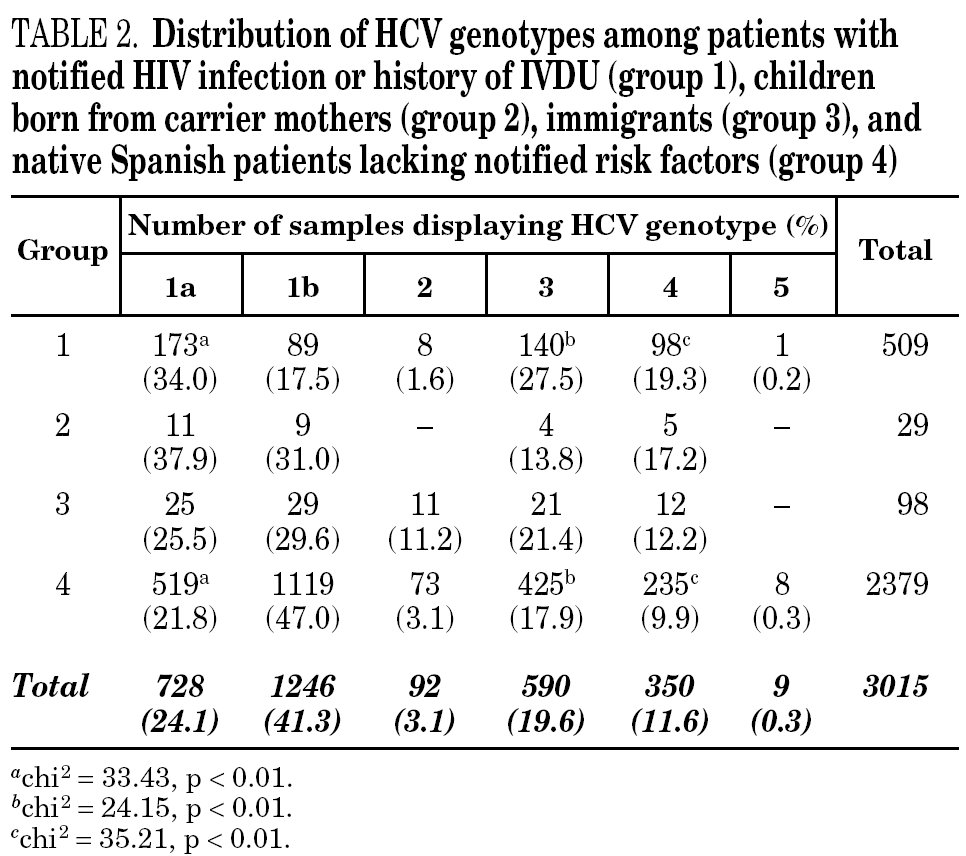
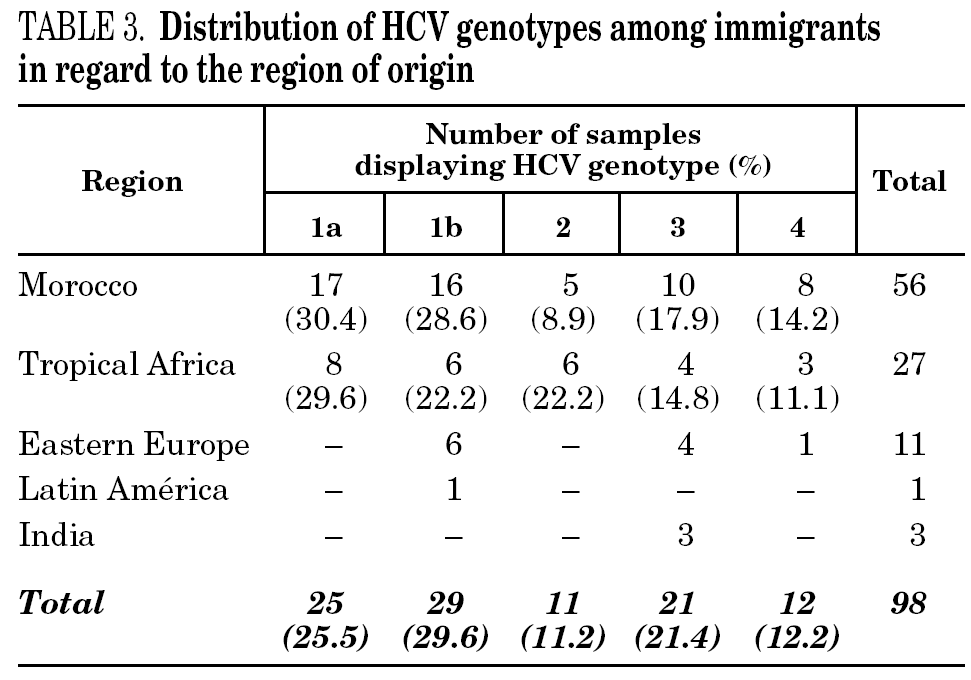
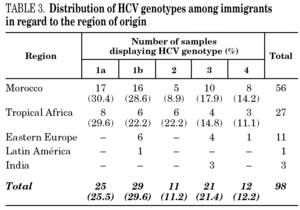
HCV genotype 1 strains present in 146 samples displayed hybridisation patterns that did not allow a genosubtype assignation, and were excluded from the analysis. These samples accounted for 4.6% of the total and were almost uniformly distributed along the period of the study (range: 4.1 to 4.8% per year). Table 2 summarises the results obtained on the remaining 3015 samples, according to the four groups of patients studied. Table 3 details those corresponding to immigrants in regard to the region of origin. Genotypes 1a, 3 and 4 were significantly more prevalent among patients from group 1 in comparison with those from group 4. Prevalence of genotype 2 was significantly higher among immigrants from the tropics of Africa in comparison with the remaining patients (22.2% vs 2.9%; chi 2 = 27.62, p < 0,01).
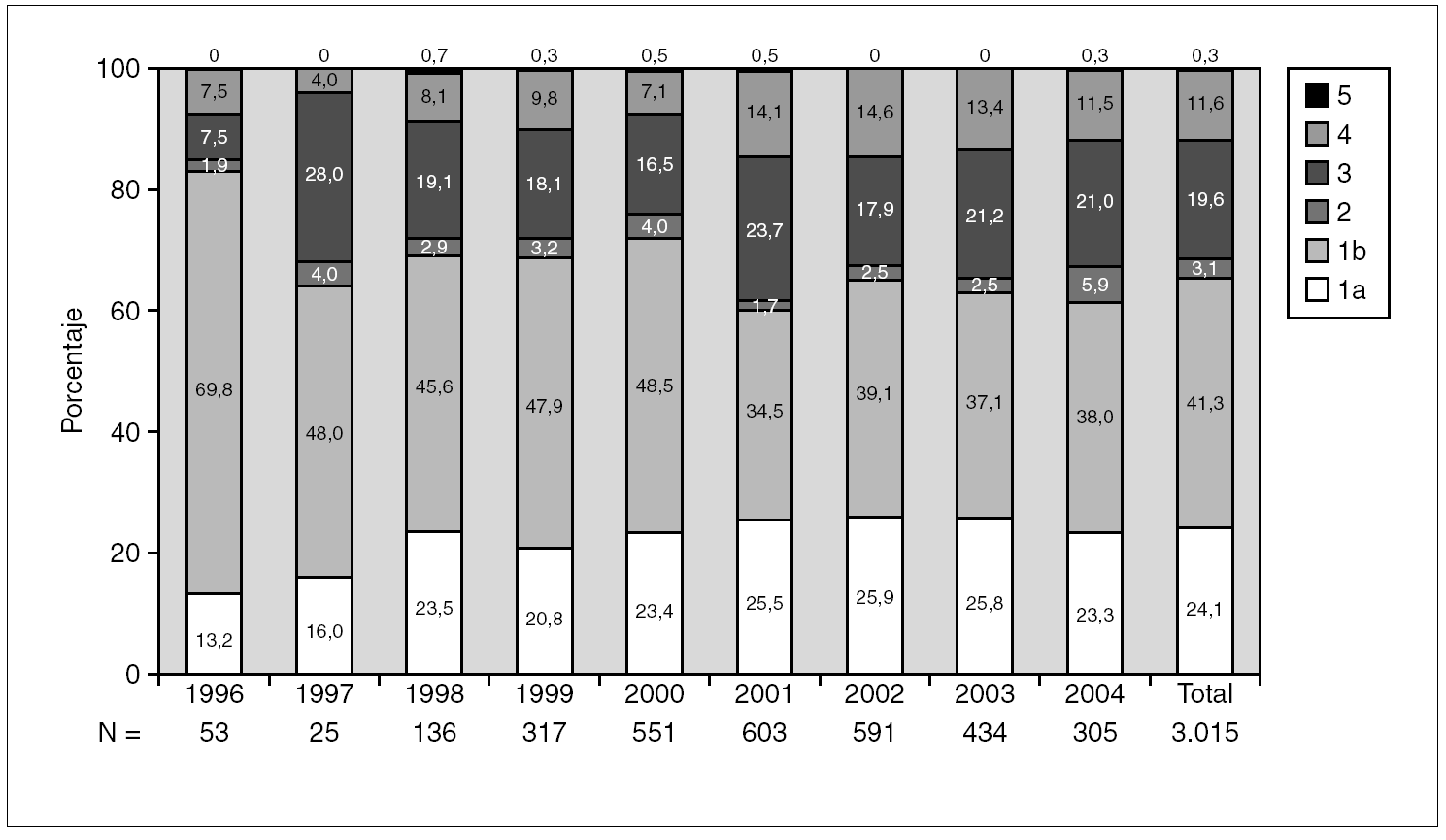
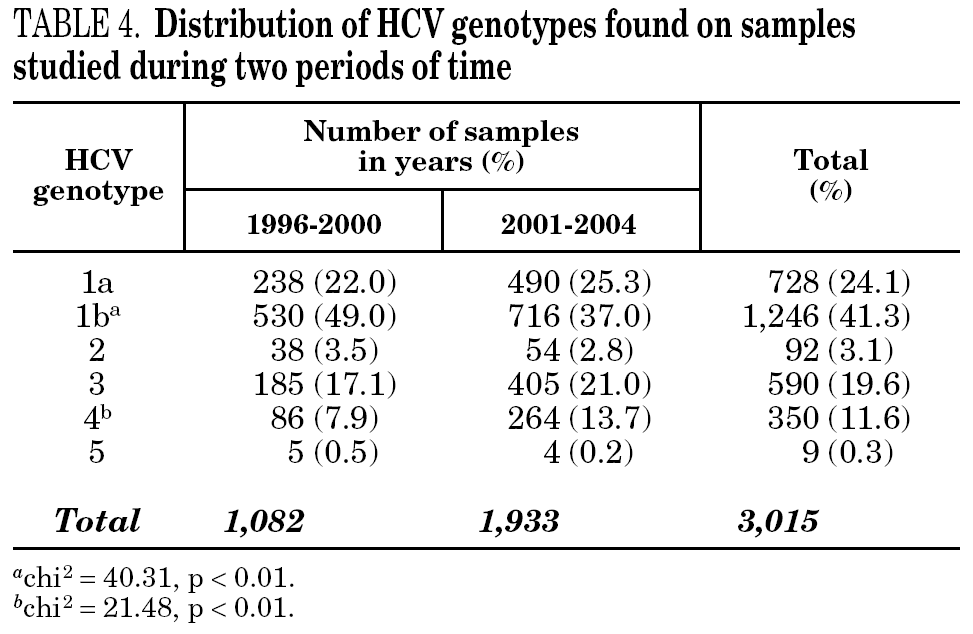
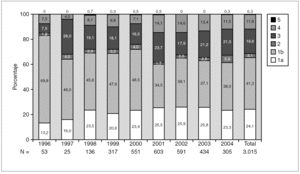
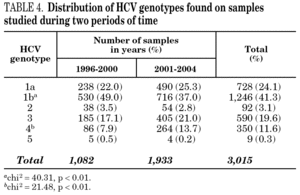
Figure 1 shows the overall prevalence of HCV genotypes on patients grouped by the year of sampling. Genotype 1b was always the more prevalent, but its overall prevalence was just 41.3%. Since a increasing trend was observed for the overall prevalence of genotypes 1a and 4, a comparison was done with samples grouped into two periods of time (1996-2000 and 2001-2004) (table 4). A significant increase of the prevalence of genotype 4 was found, together with a decrease of the prevalence of genotype 1b.
Figure 1. Prevalence of HCV genotypes among 3,015 samples from unselected carriers from Spain, grouped by time of sampling.
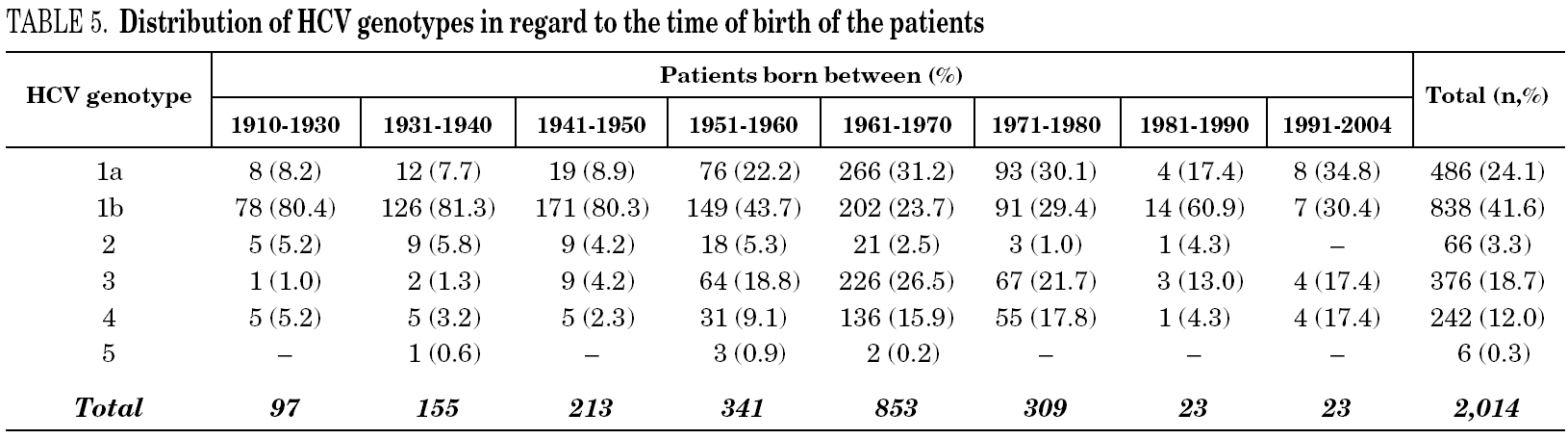
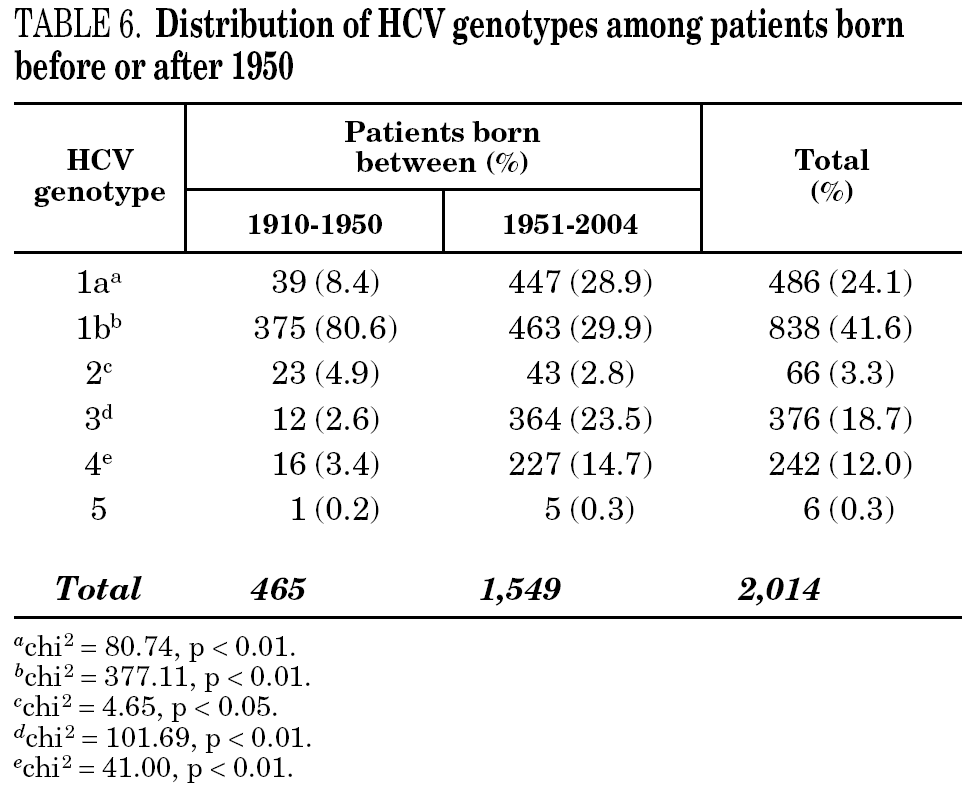
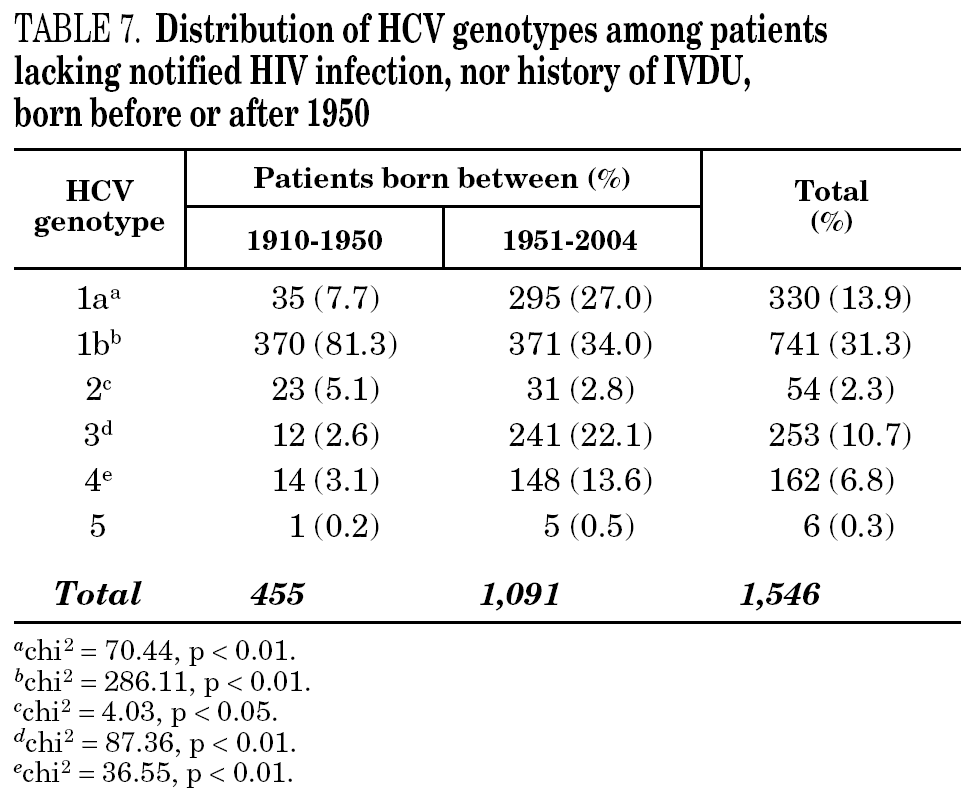
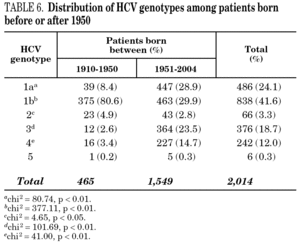
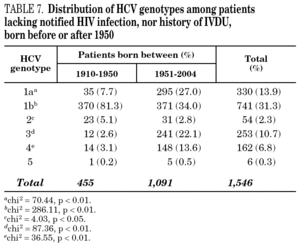
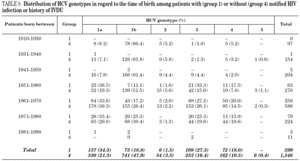
Table 5 shows the prevalence of HCV genotypes in regard to the year of birth of the patients. Genotype 1b was significantly more prevalent among those born before 1951, and the prevalence of genotypes 1a, 3 and 4 was significantly higher among patients born after 1950 (table 6). Similar findings in regard to age were found among adult patients lacking notified HIV infection and history of IVDU (tables 7 and 8). Among the latter, a significant switch-up of the prevalence of genotypes 1a and 3 was observed among patients born in 1951-1960 in comparison with those born before 1951 (7.7% vs. 19.3%, chi 2 = 20.39, p < 0.01 for genotype 1a; 2.6% vs. 15.6%, chi 2 = 39.17, p < 0.01 for genotype 3). For genotype 4, such switch-up in prevalence was, however, delayed to patients born in 1961-1970 (4.6% vs. 14.5%, chi 2 = 37.99, p < 0.01).
Discussion
Data regarding prevalence of HCV genotypes in Europe suggest that genotype 3 produced a recent epidemic of HCV infection among the injection drug users and spread subsequently to other population groups that had been already penetrated by genotype 1 strains, mainly by genotype 1b, in the past 2,12-14.
The present investigation is affected by limitations that are characteristic of retrospective studies, such as the presence of uncontrolled sampling variations from year to year, the lack of representativity of the sample in terms of population, or the risk of underreporting of relevant epidemiological data for the patients included. With these limitations in mind, the results show that Spanish HCV carriers born before 1951 are mainly infected by genotype 1b. In addition, they show that genotype 1a and 3 strains are significantly more frequent among patients born after 1950, and confirm the high prevalence of these genotypes among patients displaying significant risk factors, such as HIV co-infection or history of IVDU. Since the acquisition of the HCV infection is very uncommon before the age of 20, this is consistent with the introduction, 20-30 years ago, of genotype 1a and 3 strains into the IVDU population and with its spread to other population groups. Evidence for such introduction of HCV genotype 1a together with genotype 3 has been also reported from other European countries 12-14.
In agreement with prior findings 9-11, the age-specific patterns of prevalence found for genotype 4 indicate that this HCV genotype has been also introduced recently in Spain. They suggest, in addition, that the import of this genotype was independent of the import of genotypes 1a and 3 and happened around 10 years later. In support of this interpretation, an increasing prevalence of genotype 4 among the youngest HCV carriers have been reported from Italy 15-17, and a significant association of this genotype with infections acquired after 1990 has been found in Austria 18.
It seems very likely that introduction of genotype 4 into Europe and the United States has been related with injection drug use 15,19,20, and the findings of this investigation agree with this proposal. The significant increase in prevalence of genotype 4 found among patients without notified high risk factors born after 1960 suggest, in addition, that this genotype begun to spread to other population groups at the beginning of the 80's decade.
The diversification observed for genotypes 1 and 2 in Africa 21 and the geographical distribution found for genotypes 4 and 5 lead to suggest an African origin for all these four HCV genotypes. Import of African strains of hepatitis B virus (HBV) into Spain has been recently documented by the finding of genotype A/ayw1 and genotype E strains among native Spanish carriers 22,23. It seems, therefore, likely that introduction of African HBV and HCV strains into the Spanish population have been epidemiologically related. It is thought that the import of HCV genotype 3 and HBV genotype D/ayw3 strains original from India into the European population of IVDU were also two epidemiologically related events.
Acknowledgments
The authors wish to thank Teresa Arias, Consuelo Elola, José A. López and Inés Parera for their excellent technical assistance, as well as the following health care institutions, which sent the samples used in the study: Hospital de la Inmaculada, Hospital Comarcal de Ronda, Hospital Comarcal de La Línea de la Concepción, Hospital Infanta Helena, Hospital San Juan de la Cruz, Hospital de San Agustín, Hospital de Poniente, Hospital Virgen de las Nieves, Hospital de la Axarquía and Centro Regional de Transfusión Sanguínea de Málaga, Andalucía; Hospital San Jorge and Hospital Miguel Servet, Aragón; Hospital de San Agustín, Asturias; Hospital Son Dureta, Hospital Virgen de Montetoro and Policlínica Miramar, Baleares; Hospital Virgen de Candelaria, Canarias; Hospital Marqués de Valdecilla, Hospital Comarcal Sierrallana and Banco de Sangre de Cantabria, Cantabria; Hospital General Universitario de Guadalajara, Hospital de Santa Bárbara and Hospital Comarcal de Hellín, Castilla-La Mancha; Hospital del Bierzo, Hospital General de Segovia and Hospital General de Soria, Castilla y León; Hospital Comarcal de Palamós, Hospital Comarcal de Figueres, Laboratori de Referència de Catalunya and Clínica Renal Rotellar, Cataluña; Centro de Donación de Sangre de la Cruz Roja, Ceuta; Hospital Ciudad de Coria, Hospital Virgen del Puerto, Hospital Comarcal de Llerena, Hospital Campo Arañuelo and Complejo Hospitalario de Cáceres, Extremadura; Hospital da Costa, Complejo Hospitalario de Pontevedra and Hospital de Valdeorras, Galicia; Fundación Hospital de Alcorcón, Hospital de La Princesa, Hospital La Paz, Hospital de Getafe, Hospital Gregorio Marañón, Hospital Niño Jesús, Hospital Gómez Ulla, Fundación Jiménez Díaz, SaniLab, Banco de Sangre de la Cruz Roja and Centro de Transfusión de la Comunidad de Madrid; Hospital Virgen de la Arrixaca, Murcia; Hospital de Navarra; Banco de Sangre de Guipúzcoa, País Vasco; Hospital General de Elche, Hospital Comarcal de San Juan, Hospital Comarcal de Villajoyosa, Hospital General de Alicante and Hospital General de Castellón, Valencia.
Correspondencia: Dr. J.M. Echevarría.
Servicio de Microbiología Diagnóstica.
Centro Nacional de Microbiología.
Instituto de Salud Carlos III.
Ctra. Majadahonda-Pozuelo, km 2.
28220 Majadahonda. Madrid. Spain.
Correo electrónico: jmecheva@isciii.es
Manuscrito recibido el 27-10-2004; aceptado el 29-4-2005.