It has been reported in several countries that Chlamydia trachomatis genotypes D, E, and F are the ones more frequently associated with urogenital infections. In Mexico, the prevalence of serovars and genotypes is unknown.
Material and MethodsOne hundred and fifty-two endocervical swabs were collected from infertile women to test for C. trachomatis. The PCR-based RFLP and automated-sequencing methods of ompA gene was used to identify the C. trachomatis genotypes. Sequences of 891 pb obtained were aligned with currently available chlamydial sequences from GenBank to identify the corresponding genotype.
ResultsTwenty-four women with infertility (15.8%) were positive for C. trachomatis. According to the RFLP and nucleotide sequences results the most prevalent ompA genotype corresponded to serovar F (n=13 [54.2%]), followed by serovars E (n=2 [8.7%]), G (n=2 [8.7%]), K (n=2 [8.7%]) and LGV (n=2 [8.7%]), while serovars D, H and Ia were less prevalent (all n=1 [4.2%]). None of the patients who were positive to genovar L2 had symptoms of lymphogranuloma venereum (LGV). Nucleotide sequences analysis showed a new genovariant of L2, which was different to L2b to L2f. Mutation points were observed in VS1 domain of Omp A.
ConclusionsIn this study the most common genotypes were F. Furthermore, the L2 genovariants were demonstrated in infertile women without signs and symptoms of LGV disease. Presence of point mutations in L2 genotype sequences were seen by which there is a need for further research in order to identify new L2 genetic variants that exist in Latin America.
En diversos países se ha informado que los genotipos de Chlamydia trachomatis más frecuentes y que están asociados a infecciones urogenitales son: D, E y F. En México, la prevalencia de los serotipos y genotipos no se conoce.
Material y métodosSe obtuvieron ciento cincuenta y dos hisopos de muestras endocervicales de mujeres infértiles para detectar C. trachomatis. La identificación de los genotipos de C. trachomatis se realizó mediante la técnica de PCR basado en RFLP y en el método automatizado de secuenciación para el gen ompA. Para identificar el genotipo, la secuencia de 891 pb obtenida se alineó con secuencias del gen ompA de Chlamydia disponibles en el GenBank.
ResultadosVeinticuatro mujeres con infertilidad (15,8%) fueron positivas para C. trachomatis. De acuerdo con los métodos de RFLP y de secuencición de nucleótidos, el genotipo más frecuente correspondió al serotipo F (n=13 [54,2%]), seguida por el serotipo E (n=2 [8,7%]), G (n=2 [8,7%]), K (n=2 [8,7%]) y L2 (n=2 [8,7%]), mientras que los serotipos D, H e Ia fueron menos frecuentes (todos, n=1 [4,2%]). Ninguno de los pacientes con resultado positivo para el serotipo L2 tuvo síntomas de linfogranuloma venéreo (LGV). El análisis de las secuencias nucleotídicas del serotipo L2 mostró una nueva genovariante diferente de la L2b a la L2F. Los puntos de mutación en esta nueva genovariante se observaron en el dominio VS1 del gen ompA.
ConclusionesEn este estudio el genotipo más frecuente fue el F. Además, se demostró la presencia del genotipo L2 en mujeres infértiles que no mostraron signos y síntomas de la enfermedad de LGV. Se evidenciaron mutaciones puntuales en las secuencias nucleotídicas del genotipo L2 por lo cual hay necesidad de una mayor investigación para identificar la existencia de nuevas genovariantes L2 en América Latina.
Chlamydiae are intracellular bacterial pathogens that cause a spectrum of clinically significant diseases in humans, and Chlamydia infections are highly prevalent sexually transmitted bacterial infections.1 According to the World Health Organization, 92 million infections with Chlamydiae are detected globally each year.2 Complications caused by Chlamydial infection are disproportionately suffered by women, 70% of whom are asymptomatic; if they do not receive appropriate treatment they may develop pelvic inflammatory disease (PID) followed by ectopic pregnancy, tubal infertility or chronic pelvic pain and subsequent scarring of the Fallopian tubes.1,2
The Major Outer Membrane Protein (MOMP) constitutes approximately 60% of the proteins of the outer membrane of Chlamydia trachomatis; it is coded by the ompA gene which contains four variable segments or variable domains (VD) separated by five highly conserved segments.3 Three of these VD segments are exposed surfaces, enabling the serovars to be classified.3,4 Currently, there are 19 serovars recognized for C. trachomatis, all with well-established profiles.4
In several countries, sexually transmitted genotypes of C. trachomatis- D, E, F, G, H, I, J and K produce cervicitis while L1, L2 and L3 are associated with the illness known as lymphogranuloma venereum (LGV).1 In developed countries, it has been reported that genotypes D, E, and F are more commonly associated with cervical, vaginal and urethral infections.5,6 In the majority of Latin American countries the most frequent genotypes are unknown. Brazil and Argentina have reported that these same genotypes are common in patients with endocervical infection.7,8 In Mexico, the diagnosis of Chlamydial infection is not routine and infection is not notified to the Health Department. However, studies conducted in Mexico regarding the diagnosis of Chlamydial infections have shown infection rates varying between 4% and 28% of sexually mature women.9–11 This wide range could be due to the different techniques used for the detection of Chlamydia, as well as to the different population types. The significance that this pathogen has for the Mexican population seems clear. Although several studies have established the prevalence of Chlamydial infection in Mexico, the prevalence of different serovars and genotypes is unknown. This study describes the prevalence of C. trachomatis genotypes detected in infertile women being treated at the National Perinatology Institute.
Materials and methodsClinical SamplesA total of 152 endocervical swabs were collected from infertile women between the ages of 20 and 35 years who were receiving treatment at the National Perinatology Institute (INPer) and by written informed consent of the use of their endocervical sample for Chlamydial research. The swabs were placed in 2SP medium and then stored at −20°C until analysis.
DNA extractionDNA was isolated using a standard protocol employing proteinase K digestion, phenol-chloroform-isoamyl extraction and ethanol precipitation, as was described previously.12
PCR of endocervical samplesC. trachomatis was detected by PCR to amplify a sequence of Omp A gene which generated a fragment of about 1142bp. The primers used to amplify ompA gene were those reported by Yang et al.11; (OMP1 [GCC GCT TTG AGT TCT GCT TCC TC <] and OMP2 [ATT TAC GTG AGC AGC TCT CTC AT]). PCR was performed with 2.7mM MgCl2, 0.2mM dNTPs, 30 pM of each primer, 2.5 U of Taq polymerase (GoTaq® Flexi DNA Polymerase Promega© USA) and 5μl of the DNA sample in a final volume of 25μl. The reaction mixture was incubated for 5minutes at 95°C, followed by 35 cycles of 1minute at 95°C for denaturation, 1minute at 59°C for annealing, and 1minute at 70°C for extension, and a final elongation step of 5minutes at 70°C in a thermal cycler (Programmable Thermal Controller PTC-100 MJResearch© USA). The secondary PCR reaction contained 2μl of the initial PCR product, which generated a fragment of 879bp; the primers used were P3 (T GAC TTT GTT TTC GAC CGT GTT TT) and P4 (TTT TCT AGA TTT CAT CTT GTT CAA T/CTG) also described by Yang et al13. Thirty-five amplification cycles were carried out; each consisted of one minute at 95°C, one minute at 59°C and one minute at 70°C. The products from the initial PCR and secondary PCR were visualized by ethidium bromide staining on 2% agarose gel after electrophoresis. C. trachomatis serovar L2 (L2 434/Bu) DNA was used as a positive control.
RFLP analysisSecondary PCR products were submitted to restriction endonuclease AluI digestion (Invitrogen) of ompA gene for ten hours at 37°C; 20μl of reaction mixture contained 2 U of restriction endonuclease AluI, 8μl of PCR products and 2μl of restriction endonuclease buffer. Restriction profiles were analyzed by 10% polyacrylamide gel electrophoresis at 8V/cm for two hours.
Sequencing of the ompA geneThe sequencing of the ompA gene was carried out on an ABI PRISM 310 genetic analyzer (PE Biosystems) using a BigDye DNA sequencing kit (PE Biosystems) according to the manufacturer's instructions. P3 and P4 primers were used to cover approximately 879 pb of ompA gene of C. trachomatis.
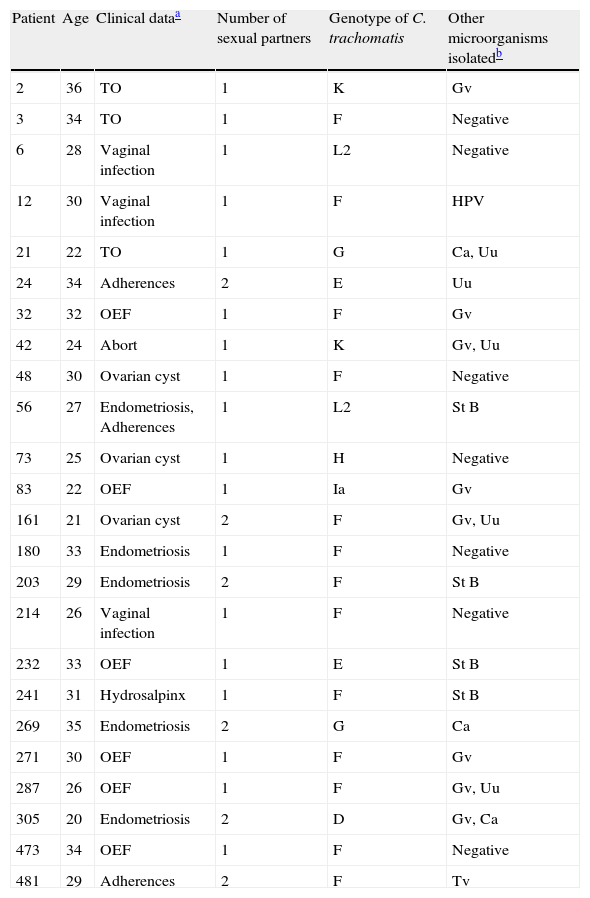
ResultsA set of 152 endocervical samples from infertile women (who had been infertile for approximately three years) were analyzed to investigate C. trachomatis infection. Women were aged between 21 and 35 years with a mean age of 27.5. They were all married and reported having an active sex life and did not use any type of contraception method. All patients were asymptomatic to PID or cervical secretions (except for samples 6, 12 and 214). Out of the 152 samples, 24 (15.8%) tested positive for C. trachomatis and 128 were negative. The positive samples were subjected to a Nested- PCR (secondary PCR) for ompA gene amplification, RFLP analysis and nucleotide sequences. According to the RLFP-PCR results, the most prevalent ompA genotype corresponded to serovar F (n=13 [54.2%]), followed by serovars E (n=2 [8.7%]), G (n=2 [8.7%]), K (n=2 [8.7%]), and L2 (n=2 [8.7%]), while serovars D, H, and Ia were less prevalent (all n=1 [4.5%]) (Table 1). The sequencing of 879 pb amplicon obtained confirmed these serovars (Fig. 1).
Genotypes of Chlamydia trachomatis detected in endocervix from Mexican women with infertility.
| Patient | Age | Clinical dataa | Number of sexual partners | Genotype of C. trachomatis | Other microorganisms isolatedb |
| 2 | 36 | TO | 1 | K | Gv |
| 3 | 34 | TO | 1 | F | Negative |
| 6 | 28 | Vaginal infection | 1 | L2 | Negative |
| 12 | 30 | Vaginal infection | 1 | F | HPV |
| 21 | 22 | TO | 1 | G | Ca, Uu |
| 24 | 34 | Adherences | 2 | E | Uu |
| 32 | 32 | OEF | 1 | F | Gv |
| 42 | 24 | Abort | 1 | K | Gv, Uu |
| 48 | 30 | Ovarian cyst | 1 | F | Negative |
| 56 | 27 | Endometriosis, Adherences | 1 | L2 | St B |
| 73 | 25 | Ovarian cyst | 1 | H | Negative |
| 83 | 22 | OEF | 1 | Ia | Gv |
| 161 | 21 | Ovarian cyst | 2 | F | Gv, Uu |
| 180 | 33 | Endometriosis | 1 | F | Negative |
| 203 | 29 | Endometriosis | 2 | F | St B |
| 214 | 26 | Vaginal infection | 1 | F | Negative |
| 232 | 33 | OEF | 1 | E | St B |
| 241 | 31 | Hydrosalpinx | 1 | F | St B |
| 269 | 35 | Endometriosis | 2 | G | Ca |
| 271 | 30 | OEF | 1 | F | Gv |
| 287 | 26 | OEF | 1 | F | Gv, Uu |
| 305 | 20 | Endometriosis | 2 | D | Gv, Ca |
| 473 | 34 | OEF | 1 | F | Negative |
| 481 | 29 | Adherences | 2 | F | Tv |
Phylogenetic tree analyses were used to demonstrate the evolutionary relationships between clinical isolates from infertile women and reference strains of Chlamydia trachomatis obtained from GenBank. Nucleotide sequences of the ompA gene determined in this study were aligned using the MEGA program (version 4).
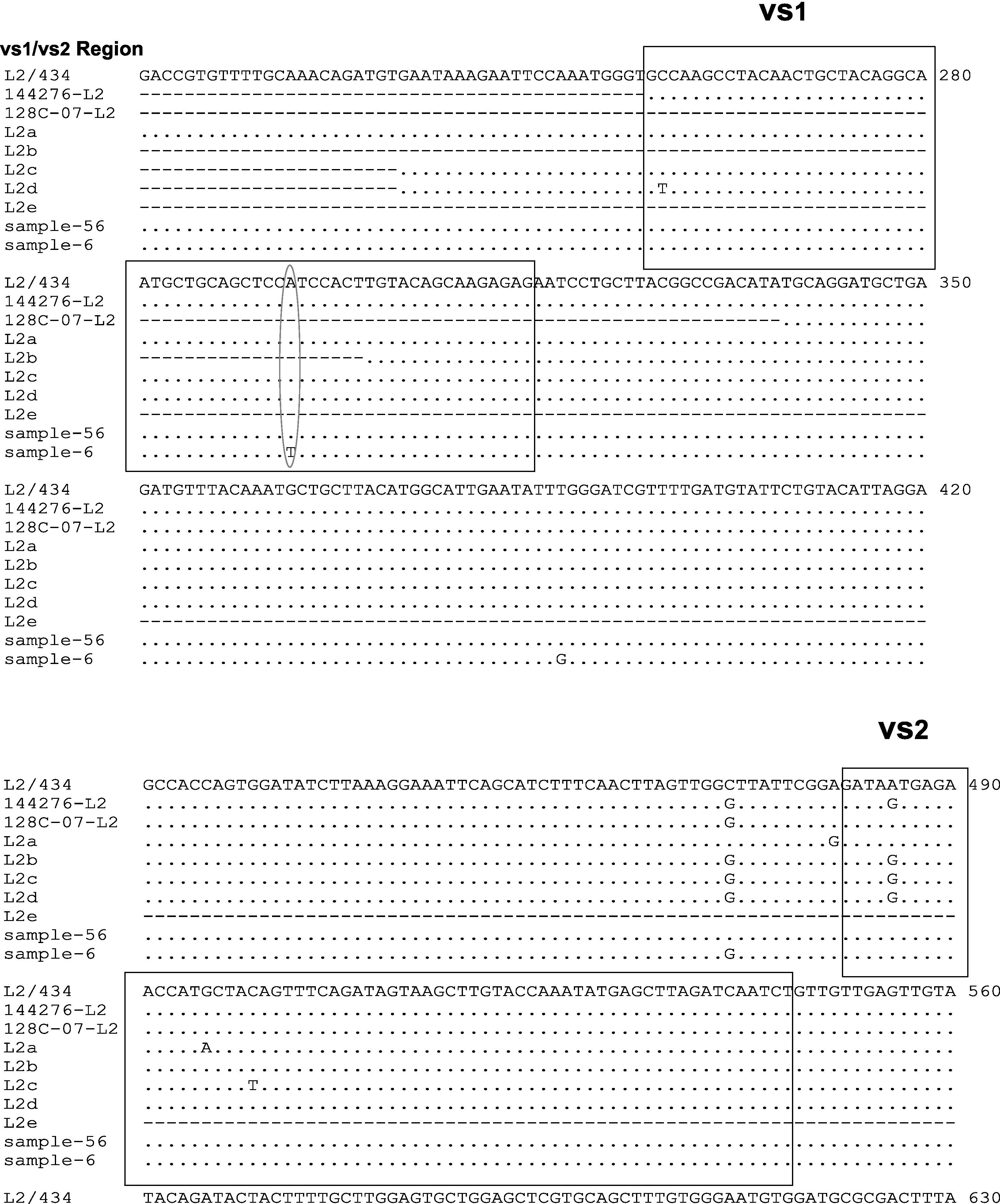
Two samples had a similar nucleotide sequence to L2/434 genotype. The 56 sample did not show any point mutation while the 6th sample had two point mutations in variability regions, one of them in vs1 in the 294 nucleotide (A294T), and other in vs4 in the 1012 nucleotide (C1012T). Another point mutation at 388 nucleotide (T388G) was observed, which belonged to the constant region of gene ompA (Fig. 2). The amino acid sequences analyses in these mutation points showed changes at Trp60Gly (A294T) and Leu268Phe (C1012T), Fig. 2.
Nucleotide sequence analysis of vs1 and vs4 domains of ompA gene from L2 genovariants. Comparison of nucleotide sequences of strain L2/434 (GenBank no. M14738), L2/144276 (GenBank No. DQ217607), L2/128c-07 (GenBank no. EU676181), L2a (GenBank no. AF304858), L2b (GenBank no. AY586530), L2c (GenBank no. EF460796), L2d (GenBank no. EF460797) and L2e (GenBank no. EF460798) with the L2 Mexican variants. The alignment was realized by Clustal W and Bioedit programs (version 7.0.9.0).
PCR-based RFLP analysis or sequencing of the amplified ompA gene, which encodes the MOMP, are currently considered to be more sensitive and more specific methods than serotyping for identifying C. trachomatis serovars.6 These technologies have provided valuable and sensitive means for molecular epidemiological analysis to identify high-risk groups and track sexual networks. Differentiation of chlamydia serovars in clinical isolation may be important for a thorough understanding of the pathogenesis and epidemiology of genital Chlamydial infections. The prevalence of C. trachomatis serovars has been identified in several countries of the world, with the serovars D (5-48%), D variants, E (22-44%) and F (8-20%) being predominant in urogenital infections, while G (4-7%), Ga, H (<5%), I (6%), I variants, J (5-13%), and K (5-10%), are less common.5,6,14,15 Sporadically, genital infections with serovars B and Ba also occur.14,15 In Latin America and the Caribbean, the prevalence of the serovars of C. trachomatis in endocervical and urethral infection is rare. A Brazilian study, reported by Lima et al.,7 described a prevalence of 19% in women who were treated at a public STD clinic and the genotypes associated with endocervical infections in these women were: D (33.3%), E (33.3%), F (16.7%) and K (16.7%). In a recent Argentine study, reported by Gallo et al8 it reported that most prevalent genotypes in endocervical swabs obtained were, E (46.9%), D (21%) and F (16.1%). In this study, the prevalence was different to the one reported by Lima or Gallo; the most prevalent genotype was F (54.2%), followed by genotype E (8.7%), G (8.7%), K (8.7%), and L2 (8.7%). In this study the most interesting was to identify the L2 genotype in infertile women without signs or symptoms of LGV.
LGV is a sexually transmitted disease caused by serovars L1, L2, and L3 of C. trachomatis.1 Nowadays serovar L2 is the most common cause of proctitis in Europe.16,17 LGV is endemic in Africa, India, Southeast Asia, South America, and the Caribbean.18 In this study 2/24 Chlamydia positive samples were L2 genotypes. The Mexican Secretary of Health reported that as of week 51 of year 2009, there had been 144 reported cases of LGV in Mexico, of which 84 corresponded to women and 60 to men.19 States in the Mexican Republic where women were most affected were: Baja California, Chihuahua, Sonora and Sinaloa.19 Despite evidence that women are more likely to become infected, transmission routes of LGV have not been considered. There is also a lack of socio-economic data and information on the sexual behavior of women. It is therefore necessary to conduct more studies in the future so as to enable us to identify the risk factors for acquiring this disease.
The classic picture of LGV usually involves lymph nodes and is characterized by buboes.17 However, there have been reports of unusual clinical pictures of LGV; for example, the current epidemic is mainly characterized by cases which present severe proctitis. LGV proctitis in men who have sex with men (MSM) is well recognized.17,20–22 For instance, it is still unclear why inguinal cases were less frequently found in these LGV cases. Other clinical presentations have yet to be studied in more detail since asymptomatic and sub-clinical cases were also identified.22 Only one case of urethritis due to genovar L2b had been reported so far.23 However, Gomes et al,24 recently reported the detection of 7 LGV specimens collected from men and women without symptoms or who had no clear LGV symptoms. These samples revealed ompA gene sequences different from L2/434 reference strain and from the L2b variant.24
New L2 genetic variants have been described recently, designated as L2b, L2c, L2d, L2e, L2f and L2g and exhibit mutations in the ompA gene.22,24,25 Currently, the L2b to L2e genovariant are associated with the development of proctitis in MSM, while L2f and L2g are associated with asymptomatic heterosexual men or women.22,24,25 In this study in the women who were positive to C. trachomatis L2 genotype, we analyzed the nucleotide sequence of these variants and compared them with those already reported in GenBank.
The nucleotide sequence analysis in new genovariants (L2a to L2g) reported that point mutations are observed in VS2 domain22,24,25 while in this investigation a L2 positive sample (6 strain) showed point mutations in VS1 domain. Stary and et al also describe a point mutation in VS1 in nucleotide 258 (C258T) of L2d genovariant,25 while in this study a one point mutation was observed in A294T. The difference in these point mutations may be because the LGV strains are isolated in other continents. However, one point mutation in C472G was observed in 6 samples in this study, which is very similar to the one reported in L2b, L2c, L2d, L2f and L2g genovariants; this suggest that L2 variants could be of a common origin.
Another point mutation has been reported in vs4 domain in L2e variant (C954T). In this study a point mutation in this region was observed in sample 6 in nucleotide 1012 (C1012T).
If these point mutations observed in VS1, VS2 and VS4 domains of several L2 variants provoke changes in amino acid sequence, this could be an indication that this bacterium can carry out the evasion of the immune response of host.
Although these results do not explain fully why patients infected with L2 genotype did not develop LGV symptoms, they might suggest that more than one point mutation may be present in the genome of L2 variants that do not development symptoms of LGV or proctitis. Finally, more research is needed to look for more point mutations in the genome of the new L2 genovariants identified that will enable us to explain why these genovariants do not provoke clinical manifestations of LGV and whether these could or not develop PID, tubal infertility or ectopic pregnancies.
Conflict of interestThe authors have no conflict of interest to declare.