This study analyzed the relationship between the ISEcp1 element and blaCTX-M genes of Escherichia coli isolates that produce extended-spectrum β-lactamase (ESBL) in community settings.
MethodsNineteen E. coli isolates that produced CTX-M-type β-lactamase were collected from four communities of elderly people in Shenyang, China. Polymerase chain reaction (PCR) amplification and direct sequencing were used to detect the insertion of the ISEcp1 element into the genetic environment of the blaCTX-M genes.
ResultsThe ISEcp1 element was associated with several blaCTX-M gene types, including CTX-M-14, CTX-M-24, CTX-M-22, and CTX-M-79. Sequence analysis revealed that all of the ISEcp1-like DNA sequences contained the putative promoter region that is involved in CTX-M genes transcription. ISEcp1 insertion sequences were observed 42–127bp upstream of the open reading frames (ORFs) that encode the CTX-M enzymes in all 15 strains. The CTX-M-79 β-lactamase-encoding gene was observed with a different ISEcp1 insertion site and variable sequences between the ISEcp1 and blaCTX-M-79 gene. For one strain (T298), the ISEcp1 element was disrupted by IS10.
ConclusionThis work confirmed that the ISEcp1 elements were closely linked to blaCTX-M genes in community isolates from Shenyang, China.
Este estudio analizó la asociación entre el elemento ISEcp1 y los genes blaCTX-M de los aislados de Escherichia coli que producen β-lactamasa de amplio espectro (ESBL) en marcos comunitarios.
MétodosSe tomaron 19 aislados de E. coli que producían β-lactamasa de tipo CTX-M en 4 comunidades de personas ancianas de Shenyang, China. Se utilizó la amplificación mediante la reacción en cadena de la polimerasa (PCR) y la secuenciación directa para detectar la inserción del elemento ISEcp1 en el trasfondo genético de los genes blaCTX-M.
ResultadosEl elemento ISEcp1 estuvo asociado con varios tipos de gen blaCTX-M, entre ellos CTX-M-14, CTX-M-24, CTX-M-22 y CTX-M-79. El análisis de secuencia reveló que todas las secuencias de ADN similares a ISEcp1 (ISEcp1-like) contenían la región promotora putativa implicada en la transcripción de los genes CTX-M. Las secuencias de inserción de ISEcp1 se encontraron a 42–127bp (pares de bases) aguas arriba de los marcos abiertos de lectura (ORF) que codifican las enzimas CTX-M en las 15 cepas. El gen CTX-M-79 que codifica β-lactamasa tuvo un punto distinto de inserción de ISEcp1 y secuencias variables entre ISEcp1 y el gen blaCTX-M-79. En una cepa (T298), el elemento ISEcp1 fue degradado por IS10.
ConclusiónEste trabajo confirma que los elementos ISEcp1 estuvieron estrechamente ligados a los genes blaCTX-M en aislados comunitarios de Shenyang, China.
More than 80 CTX-M enzyme variants (www.lahey.org/studies/) are currently recognized as rapidly emerging members of the clavulanic acid-inhibited Ambler class A extended-spectrum β-lactamase (ESBL) family. These enzymes are a specific concern in many areas of the world.1,2 In some countries, CTX-M-type enzymes are the most frequently isolated ESBLs from Escherichia coli.3 They have been involved in several outbreaks in long-term care facilities and are also becoming a problem in the community.4
Insertion sequences (IS), especially ISEcp1, have repeatedly been found adjacent to genes that encode CTX-M-type β-lactamase and appear to play an important role in the mobilization and expression of genes that encode these enzymes.5–8 Because the novel ESBL CTX-M-79 (GenBank accession no. EF426798) and other CTX-M types from community isolates in China are 99–100% identical to the above CTX-M lactamases, the ISEcp1 elements may be in the vicinity of CTX-M-79 and other CTX-M genes. Thus, we investigated the presence of ISEcp1 elements in association with the novel ESBL CTX-M-79 and other CTX-M types from community isolates in China.
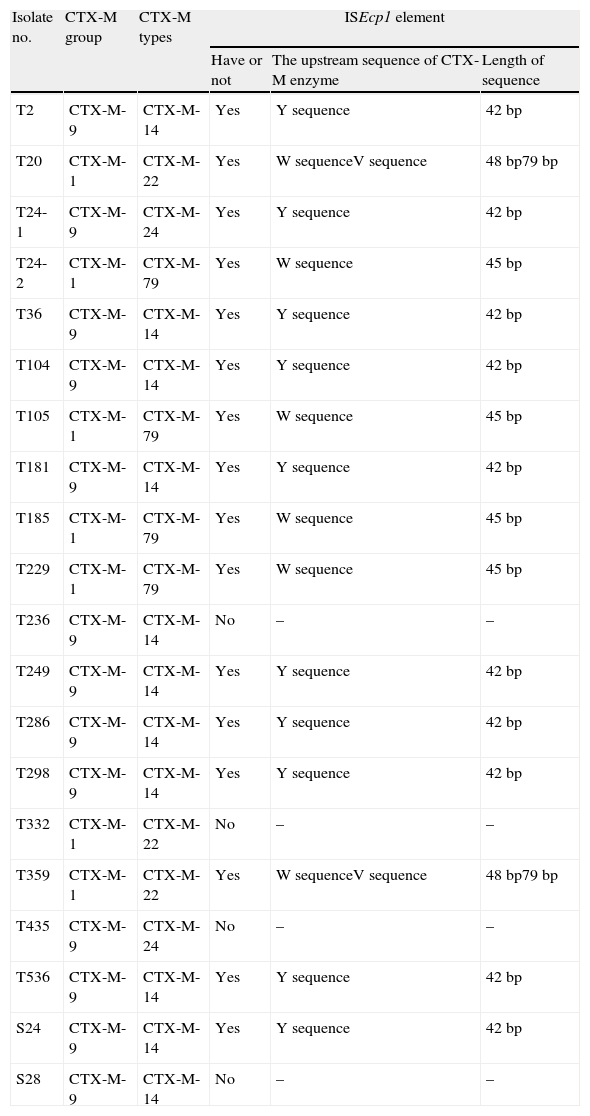
MethodsThe 19 strains of E. coli used in this study have been reported previously.9 They were isolated from rectal flora sampled from elderly people in four different communities in Shenyang, China. The strains were identified using the VITEK 2 system (bioMérieux S.A., Marcy I’Etoile, France). These strains included E. coli that produced the enzymes CTX-M-14 (11 strains), CTX-M-22 (three strains), CTX-M-79 (three strains), CTX-M-24 (one strain), and CTX-M-24 and CTX-M-79 together (one strain; Table 1). Pulsed field gel electrophoresis (PFGE) patterns from the ESBL-producers revealed a high degree of diversity.9
CTX-M types and ISEcp1 element in community isolates from Shenyang, China.
| Isolate no. | CTX-M group | CTX-M types | ISEcp1 element | ||
| Have or not | The upstream sequence of CTX-M enzyme | Length of sequence | |||
| T2 | CTX-M-9 | CTX-M-14 | Yes | Y sequence | 42bp |
| T20 | CTX-M-1 | CTX-M-22 | Yes | W sequenceV sequence | 48bp79bp |
| T24-1 | CTX-M-9 | CTX-M-24 | Yes | Y sequence | 42bp |
| T24-2 | CTX-M-1 | CTX-M-79 | Yes | W sequence | 45bp |
| T36 | CTX-M-9 | CTX-M-14 | Yes | Y sequence | 42bp |
| T104 | CTX-M-9 | CTX-M-14 | Yes | Y sequence | 42bp |
| T105 | CTX-M-1 | CTX-M-79 | Yes | W sequence | 45bp |
| T181 | CTX-M-9 | CTX-M-14 | Yes | Y sequence | 42bp |
| T185 | CTX-M-1 | CTX-M-79 | Yes | W sequence | 45bp |
| T229 | CTX-M-1 | CTX-M-79 | Yes | W sequence | 45bp |
| T236 | CTX-M-9 | CTX-M-14 | No | – | – |
| T249 | CTX-M-9 | CTX-M-14 | Yes | Y sequence | 42bp |
| T286 | CTX-M-9 | CTX-M-14 | Yes | Y sequence | 42bp |
| T298 | CTX-M-9 | CTX-M-14 | Yes | Y sequence | 42bp |
| T332 | CTX-M-1 | CTX-M-22 | No | – | – |
| T359 | CTX-M-1 | CTX-M-22 | Yes | W sequenceV sequence | 48bp79bp |
| T435 | CTX-M-9 | CTX-M-24 | No | – | – |
| T536 | CTX-M-9 | CTX-M-14 | Yes | Y sequence | 42bp |
| S24 | CTX-M-9 | CTX-M-14 | Yes | Y sequence | 42bp |
| S28 | CTX-M-9 | CTX-M-14 | No | – | – |
The genetic organization of the insertion sequence element ISEcp1 was investigated by polymerase chain reaction (PCR) amplification. The plasmid DNA for PCR amplification was prepared using commercial isolation kits (TIANGEN, Beijing, China) as recommended by the manufacturer. The primers used for amplification of the ISEcp1-CTX-M-1 group were the following: forward, 5′-GGA AAA CTA TCC GTA CAA GGG AGT G-3′; reverse, 5′-CCG TTT CCG CTA TTA CAA ACC-3′. The primer pairs for amplification of the ISEcp1-CTX-M-9 group were the following: forward, 5′-GGA AAA CTA TCC GTA CAA GGG AGT G-3′; reverse, 5′-GAT GAT TCT CGC CGC TGA AG-3′. Primers for the partial transposase gene of ISEcp1 were the following: forward, 5′-AAT ACT ACC TTG CTT TCT GA-3′; reverse, 5′-CAA CCA CCT TTC AAT CAT TTT T-3′. The 25μl reaction mixture consisted of 40–50ng of plasmid DNA, 2.0mM MgCl2, 200μM of each dNTP, 1×PCR buffer, 1 unit of TaKaRa TAQ DNA polymerase (Takara Shuzo Co., Ltd, Shiga, Japan) and 0.5μM of each primer (Sangon, Shanghai, China). PCR amplification was performed with a PCR System 9700 (Applied Biosystems), and the cycling conditions included a 5min initial denaturation step at 95°C, followed by 35 cycles of 1min at 95°C, 1min at 56°C and 1min at 72°C, and a final extension step at 72°C for 5min. The PCR products were purified with UNIQ-10 Column (Sangon, Shanghai, China) and sequenced on an ABI PRISM 3730 sequencer (Applied Biosystems) in both directions using the dideoxy chain termination method. The nucleotide sequences and deduced protein sequences were analyzed using BLAST and BioEdit software.
The nucleotide sequence data reported in this paper are available in the GenBank nucleotide database under accession no. FJ169498.
ResultsThe insertion sequence ISEcp1 was identified upstream of the blaCTX-M gene in 15 strains using PCR, but was not observed in four isolates: T236, T332, T435 and S28 (Table 1). The length of the PCR fragment amplified for the partial transposase gene of ISEcp1 was 0.5kb. The PCR products amplified for the ISEcp1-CTX-M group were 1.2kb, with the exception of one strain T298. For T298, the PCR fragment was approximately 2.5kb, suggesting the insertion of additional DNA. Direct sequencing of the PCR products identified the insertion of an IS10 sequence within ISEcp1. The length of the disrupted ISEcp1 gene was 1209bp in strain T298.
The putative promoter region involved in the transcription of the blaCTX-M genes was contained within all of the ISEcp1-like DNA sequences. Sequence analysis showed that the −35 (TTGAAA) sequences near the right inverted repeat (IRR) sequences (ACGTGGAATTTAGG 14bp) at the end of the ISEcp1-like element were located 148bp upstream of the ATG site of blaCTX-M-14, and of its point mutant derivative, blaCTX-M-24, similar to what has been found in other studies (Fig. 1).5–8 In these cases, the ATG sites of blaCTX-M-22 were located 208bp downstream of the −10 (TACAAT) promoter element of the ISEcp1-like sequences, whereas the −10 (TACAAT) promoter element of the ISEcp1-like sequences was located 82bp upstream of the initiation codon in blaCTX-M-79.
ISEcp1 insertion sequences have been observed 42–127bp upstream of the ORFs that encode the CTX-M enzymes. In all 13 strains that belong to the CTX-M-9 group (CTX-M-14 or CTX-M-24), a 42bp region with an identical sequence (named Y sequence) was found upstream of the start codon of the β-lactamase gene (Fig. 1). For three strains (T20, T359, T435), the CTX-M-22 group was characterized by a 48bp region (W sequence) and the additional 79bp fragments (V sequence), which was identical to the CTX-M-3 group as described by another study.5 The CTX-M-79 cluster (assigned as the novel gene FJ169498) was characterized by a 45bp region upstream of the blaCTX-M-79 gene, which was shorter than the W sequence with the GAC nucleotide deletion according to Eckert's study (Fig. 1).5
DiscussionIn the present study, ISEcp1 was found in the 15 strains examined (15/19, 79%; Table 1), and the ISEcp1 element was associated with several blaCTX-M genes, such as CTX-M-14, CTX-M-24, CTX-M-22, and CTX-M-79 (which was first reported in Shenyang, China). A detailed analysis of the promoter regions in bacteria containing the CTX-M genes revealed that this ISEcp1 element contained the typical −35 (TTGAAA) and −10 (TACAAT) putative promoter regions, suggesting that this IS element may enhance the expression of β-lactamase genes. Previous studies have demonstrated that the ISEcp1 elements may act as key factors in the dissemination of such ESBLs.6–8 Interestingly, in one strain, T298, this ISEcp1 element was disrupted by IS10. Notably, IS10 was identified upstream of blaCTX-M-8 (AF189721) and may play a role in the mobilization of this blaCTX-M-8 gene.
Interestingly, the insertion site of this ISEcp1 element is different from the different blaCTX-M types. The distances that separate the blaCTX-M gene from ISEcp1 vary within a given cluster of these enzyme-encoding genes, suggesting that different genetic events occur.7 In the present study, a CTX-M-79 β-lactamase-encoding gene was observed with the specific insertion of ISEcp1 and different variable sequences between ISEcp1 and blaCTX-M-79 gene. Our findings suggest that the blaCTX-M-79 gene may derive from a unique origin. However, a comparison of the same blaCTX-M-type genes from different countries (France and China) revealed that the variable sequences that separate the ISEcp1 elements from the blaCTX-M genes had highly similar patterns of nucleotide sequence variation between geographic regions,5 indicating that the insertion events seem to have emerged only once for a given blaCTX-M gene and then subsequently spread throughout the world. This assumption is yet to be confirmed.
Several enterobacteria species of the Kluyvera genus are known to be natural reservoirs of CTX-M-like genes,10 but the reservoir of ISEcp1-like sequences among various human intestinal flora has not been identified.7 Future studies should look for bacterial species in animal intestinal flora, as blaCTX-M genes have also been identified in animal isolates.
Among the ESBLs, the cefotaximases (CTX-M) constitute a rapidly growing cluster of enzymes. ISEcp1 may be an efficient tool for the mobilization and expression of blaCTX-M genes.5–8 The present study clearly showed that the ISEcp1 elements were closely associated with blaCTX-M genes from community isolates in Shenyang, China. These data may shed light on the mechanism of the rapid spread of CTX-M producing strains, but our analysis has limitations. Our study investigated only 19 isolates from four communities, which may provide only limited information. Furthermore, an analysis of the downstream region of blaCTX-M genes and plasmid assays were not performed, which deserve further study. Finally, our study was conducted in our region, and the results cannot be generalized to other settings.
Conflict of interestsThe authors declare that they have no conflict of interests related to this study.
FundingThis work was financially supported by the National Natural Science Foundation of China (Grant Nos. 81101290).
We are grateful to our laboratory staff for their continued support. No declarations were made by the authors of this paper.