We performed SBT (sequence-based typing) on clinical and environmental Legionellapneumophila isolates in Shenyang (China). We analyzed and compared the results with those obtained by PFGE (pulsed field gel electrophoresis).
MethodsTwenty-two L. pneumophila isolates were collected from two patients with L. pneumophila infection, two hospitals, and 13 office buildings. There were two clinical isolates, one strain isolated from domestic tap water, another from shower water and 18 strains from cooling tower water. All these isolates were analyzed by SBT and PFGE methods.
ResultsThe 22 isolates were divided into 7 types by SBT. Five isolates belonged to novel sequence types (ST2345, ST2344, ST2406, ST2407, and ST2408) and one isolate belonged to ST328. The STs were not obtained for two of the isolates. The remaining 14 isolates belonged to ST1. PFGE typing divided the 22 isolates into 14 pulsotypes. The main pulsotype was SYC, which included seven isolates.
ConclusionBoth typing methods showed that predominant clonal lines exist in the Shenyang region, with high levels of genetic polymorphisms. Five novel STs were identified, indicating a unique genetic composition of L. pneumophila strains in this region, which are significantly different from those found in other environmental water systems in the world.
Realizamos una tipificación mediante secuenciación (SBT) de aislados clínicos y ambientales de Legionellapneumophila en Shenyang, China. Analizamos y comparamos los resultados con los obtenidos por electroforesis en campo pulsado (PFGE).
MétodosSe recogieron 22 aislados de Legionella pneumophila de dos pacientes con infección por Legionella pneumophila, dos hospitales y 13 edificios de oficinas. Hubo dos aislados clínicos, una cepa aislada del agua del grifo doméstico, otra del agua de la ducha y 18 cepas del agua de la torre de refrigeración. Todos estos aislados se analizaron por los métodos SBT y PFGE.
ResultadosLos 22 aislados se dividieron en siete tipos de SBT; cinco aislados pertenecían a nuevos tipos de secuencia (ST2345, ST2344, ST2406, ST2407 y ST2408) y un aislado pertenecía a ST328. No se obtuvieron secuenciotipos (ST) de dos de los aislados. Los 14 aislados restantes pertenecían a ST1. La tipificación por PGGE dividió los 22 aislados en 14 pulsotipos. El pulsotipo principal fue SYC, e incluyó siete aislados.
ConclusiónAmbos métodos de tipificación mostraron que existen líneas clonales predominantes en la región de Shenyang, con altos niveles de polimorfismos genéticos. Se identificaron cinco nuevos ST, lo que indica una composición genética única de las cepas de Legionellapneumophila en esta región, que difieren significativamente de las que se encuentran en otros sistemas hídricos ambientales del mundo.
Legionella pneumophila (LP) is a Gram-negative organism ubiquitously found in natural rivers, lakes, soils, and man-made water systems that include fountains, cooling towers, and bathing facilities.1 LP withstands temperatures of 50°C for several hours yet does not multiply at temperatures below 20°C.2 These organisms survive as intracellular parasites of amoebae, ciliated protozoa, or slime moulds.3 Infected amoebae are found in naturally occurring microbial communities that form biofilms.2 In humans, the disease can occur as sporadic cases or as epidemics. In the United States, nearly 8,000 people are hospitalized annually due to LP infection, most commonly pneumonia.4 LP is the most or second most frequent community-acquired pneumonia that requires admission in intensive care units. The mortality rate is higher than that due to other organisms, particularly in immunocompromised patients, and can reach 50%.5 In China, the proportion of L. pneumonia as a cause of community-acquired pneumonia (CAP) is 5.08%.6 LP pneumonia often leads to severe pneumonia. Nearly 50% of hospitalized patients with LP pneumonia need to be admitted to the ICU, with a mortality rate of 5-30%.7 Since Legionnaires’ disease is mainly transmitted via inhalation of infectious aerosols,1 it is of great epidemiological significance to perform molecular epidemiology analysis of LP isolates and to trace the source of infection.
The European Working Group for Legionella Infections (EWGLI) has established a global database of Legionella spp. infections and identified sequence-based typing (SBT) as the “gold standard” for investigations of Legionella spp. outbreaks.8 This method amplifies and sequences multiple Legionella-specific gene loci of the target isolate using polymerase chain reaction (PCR). The sequence obtained is compared with that in the SBT database established by EWGLI in order to obtain the corresponding gene locus type. Finally, the combination of all gene locus types for each isolate is used to obtain the Sequence Type (ST) of the target isolate.
In the present study, SBT was performed on seven specific gene loci (flaA, pilE, asd, mip, mompS, proA, neuA) in 22 LP isolates. The results were compared with our previous research results, that is, pulsed field gel electrophoresis (PFGE) typing results, and this provides evidence of the epidemiology of LP infection in Shenyang, China.
METHODSBacterial isolatesFrom July 2012 to September 2014, 22 LP isolates were obtained, including two respiratory clinical isolates and 20 environmental water isolates. The two clinical isolates were collected from two large hospitals in Shenyang. The hospitals have more than 6,750 operational beds and they occupy a total land area of 625,800 square meters. The isolates were gathered from different patients, and they corresponded to sporadic cases. Twenty environment isolates were collected from 13 office buildings in different areas of Shenyang. The sources of these isolates were: domestic tap water (one isolate), shower water (one isolate), and cooling tower water (18 isolates). All these isolates were plated and cultured on buffered charcoal yeast extract (BCYE) agar plates (Oxoid, Germany) for 48 h-72h at 37°C under a 5% CO2 atmosphere. The serogroup of the strains was identified by using a serum agglutination kit (Legionella antisera, Tianjin Biochip Corporatio, China). -
Experimental methodsDNA extractionGenomic template extraction kits (QIAamp DNA minikits; Qiagen, Hilden, Germany) were purchased from Qiagen (Beijing, China). DNA templates were extracted by DNA extraction kit (QIAamp DNA Mini Kits, Hilden, Germany) according to the manufacturer's instructions (https://www.qiagen.com /us/products/discovery-and-translational-research/dna-rna-purification/dna-purification/genomic-dna/qiaamp-dna-mini-kit/?clear=true#orderinginformation). Nine hundred microliters of phosphate buffer was added to a 1.5-ml centrifuge tube. One colony of every isolate was picked using an inoculating loop and added to the buffer. The tube was vortexed and centrifuged using a low-temperature high-speed benchtop centrifuge (Kendro Laboratories, USA) at 8000 xp/min for 5min. The supernatant was discarded. ATL tissue lysis buffer (provided by QlAamp DNA Mini Kit) (180μl) was added to the centrifuge tube, followed by 20μl of proteinase K. The content was mixed by repeated resuspension and vortexing, and was subsequently placed in a constant temperature water bath at 56°C for 2h. AL lysis buffer (provided by QlAamp DNA Mini Kit) (200μl) was added to the centrifuge tube and the tube was vortexed. The tube was placed in a constant temperature metal bath at 70°C for 10min, followed by the addition of 200μl of absolute ethanol and vortexing. The adsorption column (provided by QlAamp DNA Mini Kit) and collection tube (provided by QlAamp DNA Mini Kit) were assembled. All the above liquids were transferred to the tube and centrifuged at 8000 xp/min for 1min. The collection tube and the liquid were discarded and replaced with a new collection tube. Subsequently, 500μl of AW1 (provided by QlAamp DNA Mini Kit) was added to the adsorption column and centrifugation was performed at 8000 xp/min for 1min. The collection tube and the liquid were once again discarded and replaced with a new collection tube, and 500μl of AW2 (provided by QlAamp DNA Mini Kit) was added to the adsorption column. Centrifugation was performed at 13,000 xp/min for 1min. The collection tube and the liquid were once again discarded. Centrifugation was performed at 13000 xp/min for 1min and then 100μl of elution buffer AE (provided by QlAamp DNA Mini Kit) was added. The collection tube and the liquid were discarded. The collection tube was replaced with a 1.5-ml centrifuge tube. The centrifuge tube and adsorption column were left open for 5minutes to reduce DNA damage by ethanol. Centrifugation was performed again at 8000 xp/min for 1min. The liquid after centrifugation was retained and the adsorption column discarded. The resulting liquid contained the DNA of the target isolates.
Primer synthesisSpecific upstream and downstream primers targeting the flaA, pilE, asd, mip, mompS, proA, and neuA genes of LP were synthesized by Sangon Biotech (Shanghai) Co., Ltd, China. The sequences of the Legionella-specific primers are shown in the SBT database on the EWGLI website (http://www.ewgli.org).
PCRPremix Taq reaction system (30μl) comprising 15μl of Premix TaqTM (TaKaRa Bio, Japan), 1μl of upstream primer, 1μl of downstream primer, 8μl of double-distilled water, and 5μl of the extracted DNA samples were added to a 0.2-ml centrifuge tube and vortexed. The PCR reactions were performed in a Thermo Cycler (Thermo Fisher Scientific, USA). The PCR cycling parameters were as follows: denaturation at 95°C for 5min; followed by 35 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 60 s; and a final extension at 72°C for 10min.
Sequencing of PCR productsThe sequencing primers for flaA, pilE, asd, mip, proA, and neuA were identical to the respective amplification primers. The sequencing primers for mompS were mompS-450F: TTGACCATGAGTGGGATTGG and mompS1015R: CAGAAGCTG CGAAATCAG. The obtained PCR amplicons were sequenced by Sangon Biotech (Shanghai) Co., Ltd. (China).
Data analysisThe obtained gene sequences were uploaded to the SBT database on the EWGLI website (http://www.ewgli.org) for comparison, and the allele number of each gene locus was obtained. All allele numbers of the target isolates were uploaded, and the STs of the target isolates were obtained.
PFGE (Pulsed-Field Gel Electrophoresis)PFGE is based on the separation of macro-restriction fragments of the bacterial chromosome generated by digestion with an infrequent cutting site restriction endonuclease by pulse-field gel electrophoresis. In our previous study, we used the enzyme AscI as restriction endonuclease.9 Detailed PFGE experimental methods are available at http://rs.yiigle.com/CN112137201722/992083.htm
RESULTSSerogroups of the isolatesSerogroup identification using serum agglutination tests showed that the isolates belonged to LP serogroup 1 (n=20), serogroup 7 (n=1), and serogroup 8 (n=1).
PCR amplicons of specific gene lociThe sizes of the PCR amplicons of the seven specific loci were: flaA, 414bp; pilE, 460bp; asd, 576bp; mip, 559bp; mompS, 711bp; proA, 481bp; and neuA, 459bp.10 Of the 22 LP isolates, 20 had all 7 target genes amplified. The target genes asd and mip of isolates 10 and 11 were negative after repeated amplification attempts.
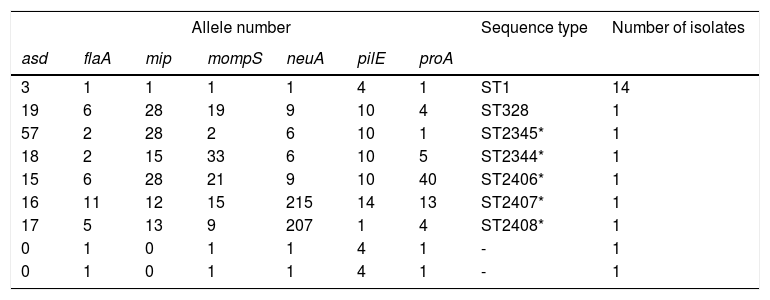
SBT resultsThe sequences were uploaded to the EWGLI database, and the allele numbers of the various housekeeping genes were obtained (Table 1). The allele numbers of the seven housekeeping genes of each target isolate were uploaded to the database to obtain the ST of the isolate. Of the 20 LP isolates with complete sequences, 14 belonged to ST1, one belonged to ST328, and the remaining five belonged to ST types that have not been reported internationally. The sequences were uploaded to the EWGLI website, with assigned ST numbers of ST2345, ST2344, ST2406, ST2407, and ST2408. The pulsotypes and STs of the 22 LP isolates are summarized in Table 1.
Description of allele number, sequence type, and number of isolates (analysis of 22 Legionella pneumophila strains).
| Allele number | Sequence type | Number of isolates | ||||||
|---|---|---|---|---|---|---|---|---|
| asd | flaA | mip | mompS | neuA | pilE | proA | ||
| 3 | 1 | 1 | 1 | 1 | 4 | 1 | ST1 | 14 |
| 19 | 6 | 28 | 19 | 9 | 10 | 4 | ST328 | 1 |
| 57 | 2 | 28 | 2 | 6 | 10 | 1 | ST2345* | 1 |
| 18 | 2 | 15 | 33 | 6 | 10 | 5 | ST2344* | 1 |
| 15 | 6 | 28 | 21 | 9 | 10 | 40 | ST2406* | 1 |
| 16 | 11 | 12 | 15 | 215 | 14 | 13 | ST2407* | 1 |
| 17 | 5 | 13 | 9 | 207 | 1 | 4 | ST2408* | 1 |
| 0 | 1 | 0 | 1 | 1 | 4 | 1 | - | 1 |
| 0 | 1 | 0 | 1 | 1 | 4 | 1 | - | 1 |
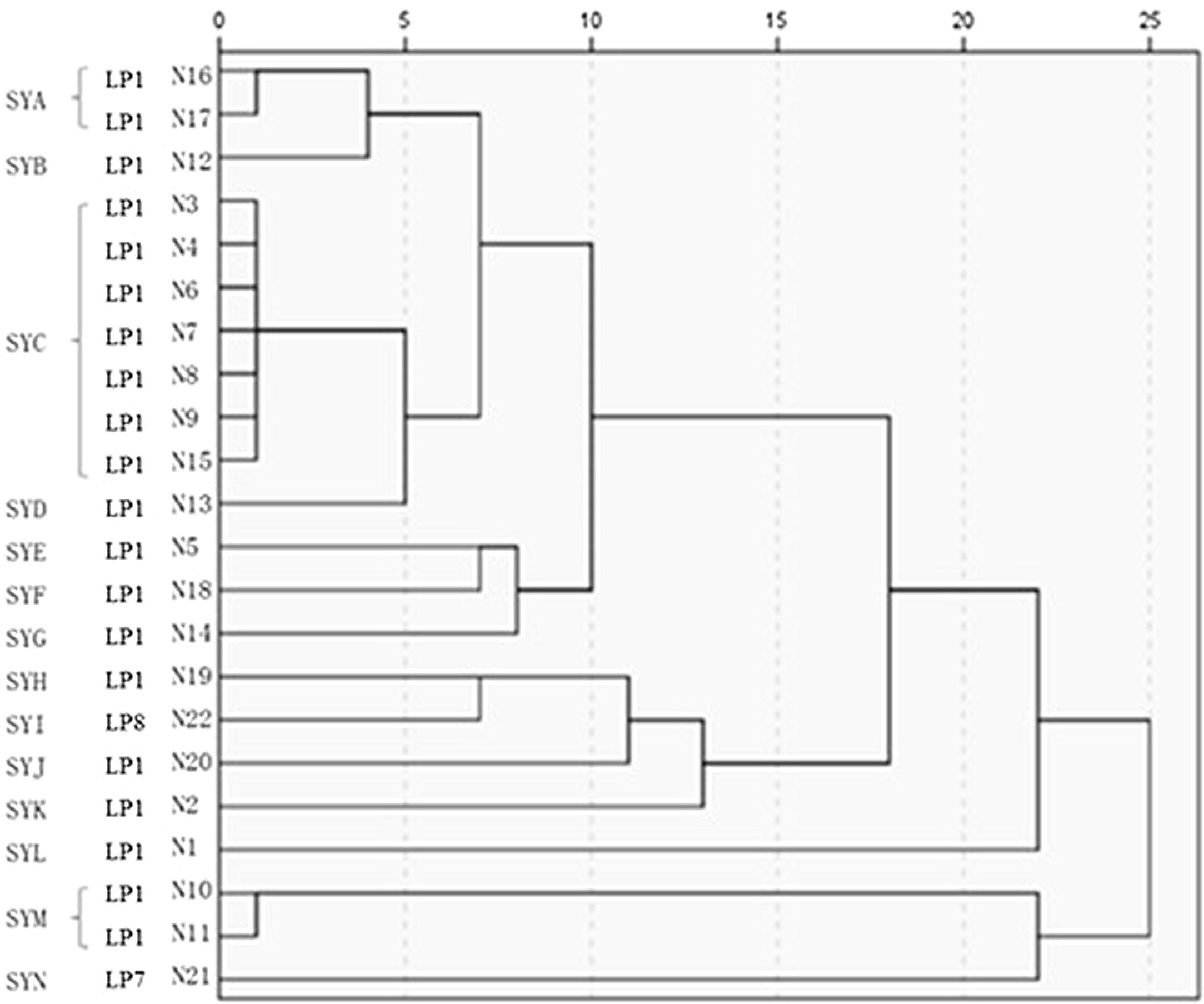
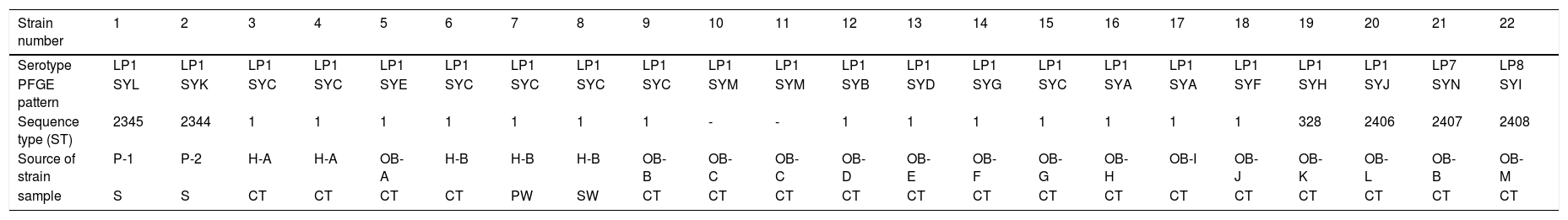
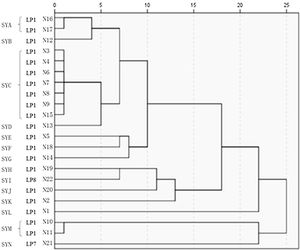
The 22 Legionella isolates in the present study had been previously tested by PFGE.11 The serogroups, PFGE types, SBT types, and sources of the isolates are summarized in Table 2. The results of cluster analysis of the 22 isolates are shown in Figure 1.
Serotypes, PFGE patterns, sequence types and source (analysis of 22 Legionella pneumophila strains).
| Strain number | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Serotype | LP1 | LP1 | LP1 | LP1 | LP1 | LP1 | LP1 | LP1 | LP1 | LP1 | LP1 | LP1 | LP1 | LP1 | LP1 | LP1 | LP1 | LP1 | LP1 | LP1 | LP7 | LP8 |
| PFGE pattern | SYL | SYK | SYC | SYC | SYE | SYC | SYC | SYC | SYC | SYM | SYM | SYB | SYD | SYG | SYC | SYA | SYA | SYF | SYH | SYJ | SYN | SYI |
| Sequence type (ST) | 2345 | 2344 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | - | - | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 328 | 2406 | 2407 | 2408 |
| Source of strain | P-1 | P-2 | H-A | H-A | OB-A | H-B | H-B | H-B | OB-B | OB-C | OB-C | OB-D | OB-E | OB- F | OB-G | OB- H | OB-I | OB-J | OB-K | OB-L | OB-B | OB-M |
| sample | S | S | CT | CT | CT | CT | PW | SW | CT | CT | CT | CT | CT | CT | CT | CT | CT | CT | CT | CT | CT | CT |
LP: Legionella pneumophila; SYA∼SYN:14 PFGE patterns; “-”: Don’t have ST type; P: patient; H: hospital; OB: office building; S: sputum; CT: cooling tower; SW: shower water; PW: pipe water;.
L. pneumophila is widely found in the environment. It can cause sporadic outbreaks through contaminated aerosols and can lead to fatal infections.12 Homologous analysis of clinical and environmental LP isolates for the determination of their genetic relationships can facilitate the tracing of the source of infection and can further the understanding of the prevalence of this organism. This knowledge is important for the prevention and control of LP infections. Current molecular typing methods used for bacterial homology analysis include PFGE, amplified fragment length polymorphism (AFLP) analysis, random amplified polymorphic DNA (RAPD) analysis, and restriction fragment length polymorphism (RFLP). However, even in a laboratory with sophisticated experimental techniques, the results of experiments conducted at different times are not 100% consistent. Disadvantages of AFLP include the imprecision of fragment sizing resulting in difficulty in comparing patterns between laboratories and platforms. For LP this method has largely been replaced by SBT.10 Since LP can cause travel-related infections, it is of considerable epidemiological significance to perform standardized molecular typing of isolates from different countries and to determine their homology.
Multilocus sequence typing was originally proposed by Enright et al.13 for the epidemiological analysis of Streptococcus pneumoniae. Gaia et al.5 applied this method to establish and evaluate the SBT method for the analysis of LP serogroup 1. The authors subsequently made improvements and performed typing using five virulence genes (flaA, pilE, mip, mompS, and proA) and one housekeeping gene (asd), and extended the application to non–serogroup 1 LP isolates. Ratzow et al.8 added a housekeeping gene (neuA) based on the data of Gaia et al.,5 which further improved the discriminatory ability of SBT. Of the seven gene loci, flaA encodes the flagellar subunit protein, pilE encodes type IV pili, asd encodes aspartate-β-semialdehyde dehydrogenase, mip encodes a protein that enhances macrophage infection, mompS encodes an outer membrane protein, proA encodes a zinc metalloproteinase, and neuA encodes an N-acyl-neuraminic cytidine transferase. EWGLI has established a standard operating procedure for SBT of Legionella spp. with accurate and objective results, and has created an online database. The database is available to the global research community and the results from each laboratory in all regions can be uploaded and compared. The SBT database for LP is constantly expanding and is important in the epidemiology of Legionella spp.5,8
Qin et al.14 conducted SBT typing on 164 LP serogroup 1 isolates from cooling towers, hot springs, and drinking water in the six cities of Shanghai, Beijing, Shenzhen, Wuxi, Jinan, and Shijiazhuang over a 7-year period. Most of the isolates belonged to ST1, accounting for 49.4% of the cases (81 isolates). ST1 existed in all cities and was the most predominant ST in China. In addition, the STs varied in different water sources and in different cities. The present study is the first to perform SBT typing of LP isolated from Shenyang. ST1 predominated (63.6%, 14 isolates) and was the main epidemic type in this region. Our findings, ST1 was the main epidemic type in this region of China, was consistent with data from other domestic studies.15–17 In addition, LP ST1 isolates of clinical relevance are spread worldwide.18 In a study performed in the United States, of clinical and environmental isolates of LP serotype 1 (LP1) from 1982 to 2012, ST1, ST35, ST36, ST37, and ST222 were responsible for both outbreak-associated and sporadic cases.19 In Germany the most common clinical ST was ST1.1 In this study, we also found ST328 in Shenyang's cooling tower water environment. This ST328 type was also isolated in hot water circuits in a large Spanish hospital.20 These environmental strains were isolated from taps and showers throughout the whole Spanish hospital.20 Besides, this ST328 environmental strain has been isolated sporadically, among others, in a nursing facility in Italy, where several cases of atypical pneumonia were recorded.21 The climate and water sources of these countries are different, so we consider that ST328 may also be widely spread globally. In addition, there were five STs that have not been previously reported in China or elsewhere. These STs were confirmed using the EWGLI website and were designated ST2345, ST2344, ST2406, ST2407, and ST2408. These results indicate that LP isolates in Shenyang display a unique gene specificity. The two LP isolates from the respiratory clinical specimens from patients belonged to novel STs (ST2345, ST2344) that have not been previously reported. Isolate 1 was collected from a 59-year-old male patient with a history of cirrhosis. Two weeks prior to disease onset, the patient received root canal treatment in a private dental clinic. The suspected route of infection was the inhalation of water contaminated with LP that was present in the tubes of the dental instruments. Isolate 2 was collected from a 45-year-old male plumber who had a history of sewer operations before the onset of the disease; the inhalation of LP present in the sewage could be a possible route of infection. These two new pathogenic STs of LP exist in the environment or in certain medical facilities in Shenyang. Isolates 20, 21, and 22 were from the environment and have not been previously reported in China and elsewhere, indicating the uniqueness of L. pneumoniae in environmental waters of Shenyang. The virulence of these three types of L. pneumoniae remains to be elucidated in future studies.
Legionella spp. can exist for a long period in low-temperature environments and in amoeba in water.22 Oberdorfer et al.23 demonstrated that this organism can also exist for a long period of time in constructed water supply systems. Therefore, long-term monitoring and genetic analyses should be performed on Legionella spp. isolated from the environment and water supply systems.
Both isolates 9 and 21 were collected from the cooling tower of the central air-conditioning system in the same office building. However, the water samples collected from different parts contained different STs, suggesting that multiple types of genetically unrelated strains could coexist in the same environment. Therefore, if an outbreak occurs, multipoint sampling should be performed in all suspicious environments for molecular analysis. For isolates 10 and 11, not all housekeeping genes could be amplified, with asd and mip genes unable to be amplified. Certain genes may differ from the housekeeping genes in the SBT database because of genetic variations in the bases within the specific primer-binding regions. It has been reported the loss of the neuA, flaA, and proA genes in LP isolates.24–26
PFGE has been widely used as the “gold standard” for the molecular typing of pathogenic bacteria,26 such as Salmonella spp., Escherichia coli, and Staphylococcus aureus. PFGE is important in the study and confirmation of the outbreaks of pathogenic bacteria.27 Zhou et al.28 optimized PFGE for LP and selected AscI as the preferred restriction endonuclease to analyze various serogroups of LP. The results indicated that clustering of PFGE patterns demonstrated high discriminatory power in typing. Despite its high discriminatory power, this method suffers some drawbacks: it is time consuming, inter-gel reproducibility is often poor, electrophoresis requires specific equipment and computer-aided imaging analysis is needed, data are difficult to exchange between laboratories making investigations of travel associated Legionella spp. disease cases more difficult.29 Despite international efforts for standardization of PFGE typing protocols (restriction endonuclease, plug preparation and electrophoretic parameters) the obtained data remain difficult to exchange. Our group has previously shown11 that the predominant PFGE type of LP in Shenyang is SYC, which includes seven molecular types isolated from four different locations and water systems, with 100% homology. Isolates belonging to serogroup LP1 can show different PFGE patterns. As shown in Table 2, the 22 isolates belonged to 14 PFGE types, which also suggests the polymorphism of LP pulsotypes in Shenyang. Isolates 1 and 2 were collected from respiratory samples of two patients with L. pneumoniae pneumonia. The two isolates were genetically unrelated and were also genetically unrelated to isolates 3 and 4 collected from the same hospital environment. In addition, the two patients had symptoms related to LP infection before admission, and both had a suspected history of exposure to a water source contaminated with LP. After admission, the initial antibiotics did not cover legionellosis, and the patients’ condition worsened. Only after adequate treatment, the patients’ condition improved. These findings suggest that the two patients were infected with LP from outside the hospital. Isolates 10 and 11 were collected from different outlets of the cooling tower in company C; their homology was 100%. Isolates 3 and 4 were collected from different outlets of the cooling tower in hospital A, and the bands were identical. These results indicate the presence of a predominant strain in the same cooling tower. Isolates 6, 7, and 8 were collected from the air-conditioning cooling tower water, drinking water, and shower water of hospital B. The three isolates belonged to the same clonal line, indicating the presence of cross-contamination of LP in different water systems of the same location. Blowing devices are often located near the cooling tower of air-conditioning system. Once LP is present in the cooling tower water system, aerosols containing the bacterium are readily formed and spread, leading to cross-contamination in different water systems. A study reported a LP in a Swedish hospital due to contamination of the shower water system.30
The present study used SBT for the typing for 22 clinical and environmental LP isolates in Shenyang. In addition, we also compared the SBT results with the PFGE results previously obtained. PFGE typing separated 14 types of patterns and SBT discriminated 7 SBT types. Both typing methods showed that predominant clonal lines exist in the region, with high levels of genetic polymorphisms. Meanwhile, five novel SBT types have been identified, indicating that the types of LP strains in this region are unique in genetic composition and differ significantly from others described from environmental water systems elsewhere in the world.
Twenty of the 22 LP isolates were assigned an ST, of which 14 isolates belonged to ST1. These 14 LP isolates displayed a certain molecular homology, but it was difficult to further analyze them by using SBT. SBT primarily distinguishes the genetic diversity of LP isolates that belong to different STs. By using this technique it is possible to compare LP isolates of different STs with the genetics of LP globally. However, it is difficult to analyze LP isolates that belong to the same ST by using this method, and its discriminatory ability is weak. The present data show that isolates of the same ST belonged to different PFGE types, suggesting that analysis by PFGE is better to SBT in tracking infections. Amemura-Maekawa et al.31 performed the typing of 31 LP1 by using SBT combined with PFGE, and found that PFGE was able to divide the isolates into 30 different types, whereas SBT could only divide the isolates into 16 types. The authors reported that the efficiency of SBT was lower than the PFGE typing method. A Japanese study divided the LP isolate ST505 into two distinct types using PFGE typing.32 Several studies utilized seven housekeeping genes to perform SBT and could not effectively trace the source of infection in hospital and community outbreaks of Legionnaires’ disease.33–35 Therefore, when SBT is unable to trace the source of infection, other methods, such as PFGE, should be used in combination. Quero et al.36 conducted an epidemiological study on 25 Legionnaires’ disease outbreaks, and concluded that the concordance between SBT and PFGE in epidemiological investigations was 100%, but their concordance in molecular typing studies was 64%.31–36 Due to the respective advantages and disadvantages of SBT and PFGE, the two methods should be used in combination when conducting epidemiological studies and tracing outbreaks due to LP.
SBT is based on the sequence comparison of several PCR-amplified DNA fragments. Since 2007, a consensus was defined and SBT is the gold standard method recommended by EWGLI for LP subtyping.37 Isolation of clinical or environmental LP strains is not an easy task. This impairs the epidemiological investigations, as both clinical and environmental isolates are required for comparisons to find the source of infection. In this respect, SBT as PCR-based methods have an advantage as they can be applied directly to DNA extracted from clinical or environmental samples without the need to isolate the strains.29 Besides, SBT is effective for epidemiological investigations on environmental and clinical isolates,38,39 and SBT has considerable advantages in the analysis of the evolutionary relationships and distribution of epidemics of bacterial strains. EWGLI has established a standard operating procedure for SBT and created an online Legionella SBT database (http://www.ewgli.org) that can be shared worldwide. SBT is simple to perform and is rapid. It has standardized procedures, and can provide objective and definitive results. In addition, the obtained isolate sequences can be compared with the data from other laboratories. Currently, SBT is widely used globally for the investigations of Legionella spp. epidemiology. Furthermore, SBT displays good discriminatory power for the typing of LP1.38,39 SBT is superior to the PFGE method for epidemiological and evolutionary analysis.29
In summary, our study indicates that LP is prevalent in public water systems and hospital water systems in Shenyang, China. SBT typing of isolates show that LP in Shenyang displays extensive genetic polymorphisms. Although some variations in SBT types have been identified, the same types of isolates still exist in some water sources. LP colonization of hospitals and public water systems may lead to LP outbreaks. The key to prevention of legionellosis is the proper maintenance of water systems in which Legionella spp. is able to grow. Active surveillance of water systems in hospitals and other facilities is required and regularly disinfection is needed.
Author contributionsLuxi Jiang, Sihong Zhao and Yu Chen designed and conducted the experiments, collected and analyzed the data, and wrote the paper. Deguang Mu, Xianghua Zhang, Jian Kang and Li Zhao designed the experiments and contributed to revising the paper. Luxi Jiang, Xu Cai, Sihong Zhao and Yu Chen conducted the experiments, collected and processed the clinical specimens.
Conflicts of InterestThe authors declare no conflict of interest.
This study was supported by grants from the National Key R & D Program of China (Grant No. 2017YFC 1309702), The National Natural Science Foundation of China (Grant No. 81170009) and Medical Science and Technology Project of Zhejiang Province (No. 2020372703) and (No. 2020KY400).