Vancomycin-resistant enterococci (VRE) among haemodialysis patients has increased rapidly and, to date, there is no report of this incidence in Portugal.
MethodsA total of 121 faecal samples were collected from haemodialysis patients, and then tested for VRE. Antimicrobial resistance, virulence and multilocus sequence typing (MLST) were studied.
ResultsVRE prevalence was 3.3%. Three VRE isolates, Enterococcus faecium, Enterococcus faecalis and Enterococcus raffinosus, were multi-resistant and vanA-positive. E. faecium and E. faecalis belonged to CC17 and CC2, respectively.
ConclusionHaemodialysis patients in Portugal are colonized with virulent, multi-resistant enterococci from high-risk clonal complexes, representing a public health concern.
La prevalencia de Enterococos Resistentes a la Vancomicina (ERV) en pacientes de hemodiálisis ha aumentado rápidamente en los últimos años. Hasta la fecha no existe ningún informe sobre esta incidencia en Portugal.
MétodosSe han tomado 121 muestras fecales de pacientes en hemodiálisis y se han analizado para la presencia de ERV. También se han tomado datos de resistencia a los antimicrobianos, virulencia y MLST.
ResultadosLa prevalencia de ERV fue del 3,3%. Tres aislamientos de ERV: Enterococcus faecium, E. faecalis y E. raffinosus, resultaron ser multirresistentes y vanA-positivos. E. faecium y E. faecalis fueron adscritos a los CC17 y CC2, respectivamente.
ConclusiónLos pacientes sometidos a hemodiálisis en Portugal son colonizados por enterococos virulentos y multirresistentes de complejos clonales de alto riesgo, lo que representa un problema de salud pública.
One of the first reports of an infection caused by a vancomycin-resistant Enterococcus (VRE) came from a chronic haemodialysis unit in England in 1988 and since then, the prevalence of VRE in patients who undergo haemodialysis has increased rapidly.1 Haemodialysis patients have played a major role in the epidemic of vancomycin resistance because this drug is commonly used in this kind of treatment.2 Given the rapid growth of the haemodialysis population, the frequency of infection and the impact of antimicrobial resistance on morbidity and mortality rates, it is of great importance to control the spread of antimicrobial-resistant pathogens among these patients.1 The recent fast spread of VRE is a global threat to public health, not only because of treatment problems, but also because of the potential for vancomycin resistance genes to spread into more virulent pathogens.3 Epidemiological data from the couple last decades have documented the emergence of enterococci as important nosocomial pathogens, exemplified by the expansion of major hospital-adapted polyclonal subclusters and paralleled by the increase in glycopeptide resistance and high-level resistance to aminoglycosides, which represent the remaining therapeutic options since penicillin resistance emerged.4 According to the latest report of the European Antimicrobial Resistance Surveillance Network, Portugal was, in 2011, the third country with the highest rate of vancomycin resistant Enterococcus faecium (20.2%) and the fourth concerning Enterococcus faecalis (3.7%); these are very high rates considering the prevalence of 1.5% for E. faecium and 0.1% for E. faecalis in the neighbour country Spain and the average vancomycin resistance prevalence of 7.2% for E. faecium and 1% for E. faecalis in the total of 29 European countries.4
To date, there is no report of VRE prevalence among haemodialysis patients from Portugal. Hence, the aim of this study was to determine VRE colonization in faecal samples of haemodialysis patients from Portugal, and also to characterize the isolates regarding resistance to other clinically important antimicrobials, virulence factors and clonality.
MethodsOne faecal sample per patient was collected from a total of 121 haemodialysis patients, between September and December 2010, from one of the largest dialysis clinics in Portugal (Nephrocare Lumiar, Lisbon). Ethical approval was obtained from the ethics committee of Nephrocare Lumiar clinic and all participants in the study provided informed consent. Each sample was transported to the laboratory in a Carry-Blair medium, seeded on Slanetz-Bartley agar plates supplemented with 4mg/mL of vancomycin and incubated for 48h at 35°C. Colonies with typical enterococcal morphology were selected and identified to the genus and species level by cultural characteristics, Gram-staining, catalase test, bile-esculin reaction and biochemical tests, using the API 20 Strep system (BioMérieux, La Palme, France). Identification was also confirmed by 16S ribosomal RNA sequence analysis. Antimicrobial susceptibility was tested by disc diffusion for 11 antimicrobials (vancomycin, teicoplanin, ampicillin, gentamicin, streptomycin, kanamycin, chloramphenicol, tetracycline, erythromycin, quinupristin–dalfopristin and ciprofloxacin) following the Clinical and Laboratory Standards Institute (CLSI) criteria.5 High-level resistance was considered for aminoglycosides. Vancomycin resistance genes, vanA, vanB, vanC1 and vanC2/3, were tested by polymerase chain reaction (PCR) in isolates with resistance or reduced susceptibility for glycopeptides.6 Macrolide [erm(A), erm(B), erm(C)], streptogramine [vat(D) and vat(E)], tetracycline [tet(M), tet(L)] and aminoglycoside [aph(3′)-IIIa, aac(6′)-aph(2″), ant(6)-Ia] resistance genes were screened by PCR in the isolates that showed resistance to these agents.6 The presence of virulence genes (ace, gelE, fsr, cpd, esp, hyl, agg and cylLLLSABM) was also studied.6E. faecium and E. faecalis isolates were characterized by multilocus sequence typing (MLST).6
Results and discussionVancomycin-resistant enterococci were detected in four of the 121 analyzed faecal samples (3.3%); this is the first report of VRE colonization rates among haemodialysis patients in Portugal. Nonetheless, VRE prevalence among dialysis patients has been evaluated before elsewhere; Kalocheretis and co-workers7 compared several studies from the USA that reported VRE frequencies ranging from 5.8% to 9.5%. In Europe, the VRE prevalence reported in Belgium was 33.2%3 and 13% in Ireland8; a much lower prevalence was reported in Greece (3.9%),7 which is similar to the results obtained in our study (3.3%). However, the use of enrichment techniques for VRE detection may have influenced the measured prevalence in the different studies.3
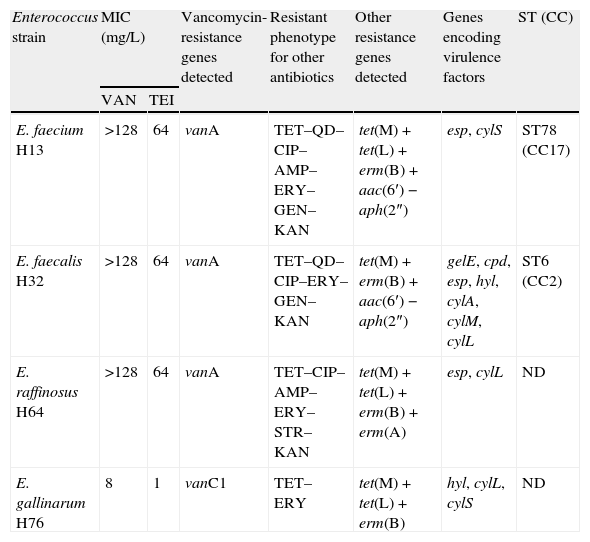
Three of the four detected VRE isolates, identified as E. faecium, E. faecalis and Enterococcus raffinosus, exhibited the acquired vanA type of resistance; the remaining isolate was identified as a naturally vancomycin resistant vanC1-harbouring Enterococcus gallinarum that carried important resistance [tet(M), tet(L), erm(B)] and virulence (hyl, cylL, cylS) genes (Table 1). All of these species cause human enterococcal infections; E. faecalis causes 80–90% and E. faecium causes the majority of the remainder. E. raffinosus is a rarely encountered species, which is often misidentified by the widely used identification systems. The presence of the vanA gene is of clinical relevance, because it is generally located on mobile genetic elements and already proved its capability to be transferred to other more virulent and potentially pathogenic bacteria, such as methicillin-resistant Staphylococcus aureus (MRSA).9 Furthermore, all of these vanA-containing isolates showed to be multidrug resistant, exhibiting resistance to three or more structurally unrelated antimicrobial agents of clinical relevance. Recent enterococcal population retrospective analysis suggests that the temporal evolution of the population biology of Enterococci is driven by serial epidemic waves of human specific lineages, E. faecalis ST6 and E. faecium ST78, that are emerging at a global scale in the last decade.10 The MLST analysis of the E. faecalis isolate (H32) revealed that it belongs to the CC2 (ST6), a high-risk clonal complex globally associated with hospital outbreaks and life-threatening infections.9 This major clonal lineage is dominant in the Portuguese nosocomial setting, being established in this country; these are widely disseminated multidrug resistant strains, recovered from Portuguese hospitals since 1996 but also among vanA isolates from swine manure and hospital sewage, indicating a community contamination.11 Isolate H32 showed to be highly virulent, presenting the genes coding for gelatinase (gelE), hialuronidase (hyl), sex pheromone (cpd), extracellular surface proteins (esp), and some of the genes of the cytolysin operon (cylA, cylM and cylL). According to recent studies, the diversity of resistance and virulence gene profiles showed by CC2 isolates from Portuguese extra-hospital settings supports that CC2 is particularly proficient at DNA exchange.11 The E. faecium isolate (H13) belongs to CC17 (ST78), a globally dispersed clonal lineage that is highly successful in the hospital environment and characterized by resistance to ampicillin and quinolones.9 A recent comprehensive multi-hierarchical analysis of VRE isolates from hospitals and aquatic surroundings in Portugal showed that the population structure of VRE comprises main human E. faecium lineages, ST18 being more abundant than ST78.10 Nonetheless, CC17 vanA-E. faecium are spread among Portuguese human and swine hosts12 and also among wild animals such as the Iberian Wolf13 and common buzzards.6 Portugal is one of the European countries with highest rates of VRE associated with nosocomial infections, being vanA-E. faecium isolates the main responsible.14 However, in the last decade, the prevalence of VRE E. faecium decreased from 46.6% in 2003 to 20.2% in 2011.4 Although polyclonality is frequently observed, intra- and inter-hospital dissemination of persisting E. faecium and E. faecalis clones and specific vanA transposon types seemed to have contributed to the rapid and extensive spread of VRE in Portuguese hospitals.14 Reports of a high proportion of ampicillin-resistant VRE E. faecium isolates together with MLST data suggest the wide dissemination of epidemic clones among Portuguese hospitals.14
Characteristics of vancomycin-resistant enterococci isolates recovered from haemodialysis patients in Portugal.
| Enterococcus strain | MIC (mg/L) | Vancomycin-resistance genes detected | Resistant phenotype for other antibiotics | Other resistance genes detected | Genes encoding virulence factors | ST (CC) | |
| VAN | TEI | ||||||
| E. faecium H13 | >128 | 64 | vanA | TET–QD–CIP–AMP–ERY–GEN–KAN | tet(M)+tet(L)+erm(B)+aac(6′)−aph(2″) | esp, cylS | ST78 (CC17) |
| E. faecalis H32 | >128 | 64 | vanA | TET–QD–CIP–ERY–GEN–KAN | tet(M)+erm(B)+aac(6′)−aph(2″) | gelE, cpd, esp, hyl, cylA, cylM, cylL | ST6 (CC2) |
| E. raffinosus H64 | >128 | 64 | vanA | TET–CIP–AMP–ERY–STR–KAN | tet(M)+tet(L)+erm(B)+erm(A) | esp, cylL | ND |
| E. gallinarum H76 | 8 | 1 | vanC1 | TET–ERY | tet(M)+tet(L)+erm(B) | hyl, cylL, cylS | ND |
MIC, minimal inhibitory concentration; ST, sequence type; CC, clonal complex; ND, not done; VAN, vancomycin; TEI, teicoplanin; TET, tetracycline; QD, quinupristin–dalfopristin; CIP, ciprofloxacin; AMP, ampicillin; ERY, erythromycin; STR, streptomycin (high-level); GEN, gentamicin (high-level); KAN, kanamycin (high-level).
In conclusion, the prevalence of VRE colonization in the studied haemodialysis patients was 3.3%. The majority of these enterococci possess the vanA gene and also displayed multidrug resistance to the antimicrobials used to treat severe enterococcal infections. Hence, the dissemination of these VRE is a matter of public health concern, particularly among the dialysis population, as these enterococci constitute a reservoir of resistance genes that can be disseminated to other pathogens. This study might contribute to a future understanding on the problem of VRE colonization among this risk population.
Conflicts of interestThe authors have no conflicts of interest to declare.
The authors would like to thank Dr. João Carlos Diniz, Gabriela Filipe and Jesus Salgueiro from Nephrocare Lumiar for their contribution in sample collection and also all the patients who consented for faecal sample recovery.
Susana Correia (SFRH/BD/75160/2010) and Daniela Jones-Dias (SFRH/BD/80001/2011) are supported by PhD fellowships granted by FCT (Fundação para a Ciência e a Tecnologia) and POPH/FSE (Programa Operacional Potencial Humano/Fundo Social Europeu).