Chlamydia trachomatis is one of the main etiological agents of sexually transmitted infections worldwide. In 2006, a Swedish variant of C. trachomatis (Swedish-nvCT), which has a deletion of 377bp in the plasmid, was reported. In Latin America, Swedish-nvCT infections have not been reported. We investigated the presence of Swedish-nvCT in women with infertility in Mexico.
MethodsSwedish-nvCT was searched in 69C. trachomatis positive samples from 2339 endocervical specimens. We designed PCR primers to identify the deletion in the plasmid in the ORF1, and the presence of a repeated 44bp in the ORF3. The sample with the deletion was genotyped with the genes of the major outer membrane protein A (ompA) and the polymorphic membrane protein (pmpH).
ResultsThe deletion was detected in one of the 69 samples positive C. trachomatis of 2339 endocervical exudates. The nucleotide sequence analysis of the ompA shows a high degree of similarity with the Swedish nvCT (98%), however the variant found belongs to serovar D. The nucleotide sequence of the pmpH gene associates to the variant found in the genitourinary pathotype of the Swedish-nvCT but in different clusters.
ConclusionsOur results revealed the presence of a new variant of C. trachomatis in Mexican patients. This variant found in Mexico belongs to serovar D based on the in silico analysis of the ompA and pmpH genes and differs to the Swedish-nvCT (serovars E). For these variants of C. trachomatis that have been found it is necessary to carry out a more detailed analysis, although the role of this mutation has not been demonstrated in the pathogenesis.
Chlamydia trachomatis es una de las principales bacterias que causan infecciones de transmisión sexual en todo el mundo. En 2006 se informó de una variante sueca de C. trachomatis (nvCT-sueca), que tiene una deleción de 377 bp en su plásmido. En América Latina no se ha informado de infecciones por la nvCT-sueca. El propósito de esta investigación fue la búsqueda de la nvCT-sueca en mujeres mexicanas con infertilidad.
MétodosSe analizaron 69 muestras positivas para C. trachomatis de 2.339 muestras endocervicales. Se diseñaron cebadores que identificaron la deleción de 377pb en ORF1, y detección de un tándem de 44pb repetidos en ORF3, como ocurre en la nvCT-sueca. Las muestras con la deleción fueron genotipificadas mediante los genes de la proteína principal de la membrana externa A (ompA) y de la proteína polimórfica de membrana H (pmpH).
ResultadosLa deleción se detectó en una de las 69 muestras (1,44%). El análisis de la secuencia del gen ompA mostró un alto grado de similitud con la nvCT-sueca (98%). Sin embargo, la variante encontrada perteneció al serovar D. La secuencia del gen pmpH se asoció al patotipo genitourinario, pero en diferentes clusters al de la nvCT-sueca.
ConclusionesLos resultados revelaron la presencia de una nueva variante de C. trachomatis en México con delección y que pertenece al serovar D con base al análisis in silico de los genes ompA y pmpH, y que difiere de la nvCT-sueca (serovares E). Se requiere conocer su prevalencia en México y en América Latina.
Chlamydia trachomatis is an important cause of sexually transmitted bacterial infections worldwide. Certain serovars (genovars) can lead to complications, such as pelvic inflammatory disease, ectopic pregnancy, and infertility. A variety of nucleic acid amplification tests (NAATs),1 such as nucleic acid hybridization, polymerase chain reaction (PCR) endpoint, real-time PCR, and rRNA detection have been used to diagnose C. trachomatis. Currently, the targets for the identification of C. trachomatis are sequences of major outer membrane protein (ompA) and its plasmid. In 2006, Ripa and Nilsson2 identified a deletion of 377bp in the open reading frame 1 (ORF1) plasmid sequence that led to false-negative reports by the diagnostic systems Abbott m2000® (Chicago, IL, USA) and COBAS® Amplicor/Taqman48 (Roche Diagnostics, Basel, Switzerland) in the Chlamydia diagnostic from Swedish patients. In a preliminary analysis, it was assumed that 30% of all C. trachomatis cases were caused by the Swedish-nvCT, and they belong to the serovar E, which used the Abbott or Roche system. In recent years, approximately 8000C. trachomatis cases have escaped detection.3 Worldwide, C. trachomatis cases have not diminished; instead, the number of infections has increased by 20% and continues to rise.3
Some studies have suggested that the plasmid of C. trachomatis acts as both a transcriptional regulator of chromosomal genes and a virulence factor.4 However, the effects of deletion in the plasmid in C. trachomatis strains are unknown.
In Mexico, no reports of the presence of Swedish-nvCT exist, and studies in this country have only determined the prevalence of C. trachomatis infections in the infertile population.5 The purpose of this study was to detect the presence of Swedish-nvCT in endocervical samples from Mexican infertile women. To this end, we designed a pair of primers to amplify the section of the plasmid with or without a 377bp deletion to identify Swedish-nvCT in the Mexican population.
Material and methodsDuplicate endocervical swabs were collected from 2339 infertile patients for the detection of C. trachomatis by NAATs using the COBAS® Amplicor/Taqman48 assay (Roche Diagnostics, Basel, Switzerland). All of the women were sexually active and attended the infertility clinic at the National Institute of Perinatology in Mexico City from January to December 2015 (Ethics Committee approval number: 212250-3120-10607-01-14). The inclusion of patients was reviewed by the institute's ethics committee.
Specimens were collected in Copan Universal Transport Medium (UTM-RT, Copan Diagnostics Inc., Murrieta, CA, USA). The samples were stored at −70°C before being analyzed. The second tube from the C. trachomatis-positive samples was used for total DNA extraction via the phenol-chloroform method.
The detection of C. trachomatis plasmid was carried out by with an in-house PCR assay. The test consisted of the amplification of a 241bp fragment of the ORF2 region of the cryptic plasmid. We used KL1 and KL2 primers, 5′-TCCGGAGCGAGTTACGAAGA-3′ and 5′-AATCAATGCCCGGGATTGGT-3′ respectively (IDT®, Coralville, IA, USA). PCR was performed in a 25μL reaction volume containing 10pM of each primer, 1.75μM MgCl2, 0.2μM dNTPs, 2.5U of Taq polymerase (Invitrogen, CA, USA), and 5μL of plasmid DNA from each sample. For each PCR assay, a positive control (plasmid DNA from C. trachomatis ATCC® VR-902B), a negative control (HeLa cell DNA), and a 100bp DNA ladder (Invitrogen, CA, USA) were used. Thermal cycling included an initial denaturation step at 95°C for 5min, followed by 35 cycles in a PTC-100 thermocycler (MJ Research Inc., Hercules, CA, USA). Each cycle consisted of 95°C for 45s, 58°C for 45s, and 72°C for 1min, with a final extension at 72°C for 10min. The PCR products were analyzed on 2% agarose gel in TAE buffer.
The identification of Swedish-nvCT was based on the studies of Ripa and Nilsson and Unemo et al.,2,6 we generated new oligonucleotides for the identification of Swedish-nvCT in samples. We used the consensus sequence of the National Center for Biotechnology Information (NCBI) reports of the C. trachomatis plasmid (data not shown). These primers were designed by the program Primer3plus. To identify Swedish nvCT, the oligonucleotide delC1 (5′-TTGACCACAGCGAATCTTTG-3′) targets the nucleotide sequence from position 340 to position 360, and delC2 (5′-CACAATATTGGGGGTGTTTG-3′) targets the nucleotide sequence from position 1061 to position 1081 of a cryptic plasmid from C. trachomatis. The primers (IDT®, Coralville, IA, USA) were intended to detect Swedish-nvCT, and amplification of a 742bp product reflects the presence of C. trachomatis strains with plasmids without deletions. Amplification of a 365bp product reflects the presence of strains containing plasmids with deletions. To confirm the Swedish variant, the amplification and sequencing of the ORF3 of the C. trachomatis plasmid was performed. The ctorf3 primers (5′-TTGCAGATTCATATCCAAGGAC-3′ and 5′-CCCGAGATACGATTTGTCCA-3′, Macrogen Inc., Geumcheon-gu, Seoul, Republic of Korea), were designed to detect the duplication of 44pb.
The PCRs were performed in a 25μL reaction volume. For each PCR assay, a positive control (plasmid DNA from C. trachomatis ATCC® VR-902B), a negative control (HeLa cell DNA), and a 100bp DNA ladder (Invitrogen) was used. Amplification was performed in a PTC-100 thermocycler (MJ Research Inc.). The conditions were an initial denaturation step at 95°C for 5min, followed by 30 cycles (95°C for 45s, 60°C for 45s, and 72°C for 1min), with a final extension at 72°C for 10min. The PCR products were analyzed on 2% agarose gel in TAE buffer. Subsequently, the 365bp (variant with deletion) and 595bp (ORF3) PCR's products were purified using a NucleoSpin instrument (Macherey-Nagel, Düren, Germany). The purified fragments were then frozen at −70°C until sequencing.
The Identification of the serotype of the Swedish-nvCT was realized by genotyping of the ompA gene and pmpH gene. PCR amplification and sequence analyses of endocervical samples that exhibited the presence of a plasmid with deletion were performed. Based on the work of de Jesús de Haro-Cruz et al.,7 we developed the PCR assay using the oligonucleotides OMP1 (5′-TTGACCACAGCGAATCTTTG-3′) and OMP2 (5′-CACAATATTGGGGGTGTTTG-3′) (synthesized by IDT®). The primer hybridization sites allowed us to obtain the complete ompA gene of C. trachomatis. In the case of the pmpH gene, the pmpH primers (5′-TGTTTCTTGCGGAGAAAAGG-3′ and 5′-TGAAAGAAACTTTCCCTTTACAGTT-3′, synthesized by Macrogen Inc.) were developed to obtain a product of 392 pb and where the sequence allowed us to determine the pathotype of C. trachomatis studied. The 1182bp (ompA gene) and 392bp (pmpH gene) PCR products were purified with a NucleoSpin instrument (Macherey-Nagel). The purified fragments were then frozen at −70°C until sequencing.
The PCRs were performed in a 25μL reaction volume. For each PCR assay, a positive control (total DNA from C. trachomatis ATCC® VR-902B), a negative control (HeLa cell DNA), and a 100bp DNA ladder (Invitrogen) were used. The ompA gene and pmpH gene amplification were performed in a PTC-100 thermocycler (MJ Research Inc.). The conditions were as follows: denaturation for 5min; 40 cycles of 95°C for 1min, 60°C for 1min, and 72°C for 1min; and a final extension at 72°C for 10min. The PCR products were analyzed on 2% agarose gel in TAE buffer.
The purified DNA fragments were sent to the Instituto de Biología (National Autonomous University of Mexico) for sequencing by an ABI PRISM sequencer (Applied Biosystems, Waltham, MA, USA). Reported sequences were obtained, and the ompA gene was analyzed with de Chromas software, identified by BLASTn (NCBI),8,9 and aligned with ClustalW,10 and Multalin software (version 5.4.1, Multiple sequence alignment with hierarchical clustering, France)11; its phylogenetic relationships were determined using iTOL (phylogenetic associations based on the algorithm of Fitch-Margoliashm least-squares distance methods).12
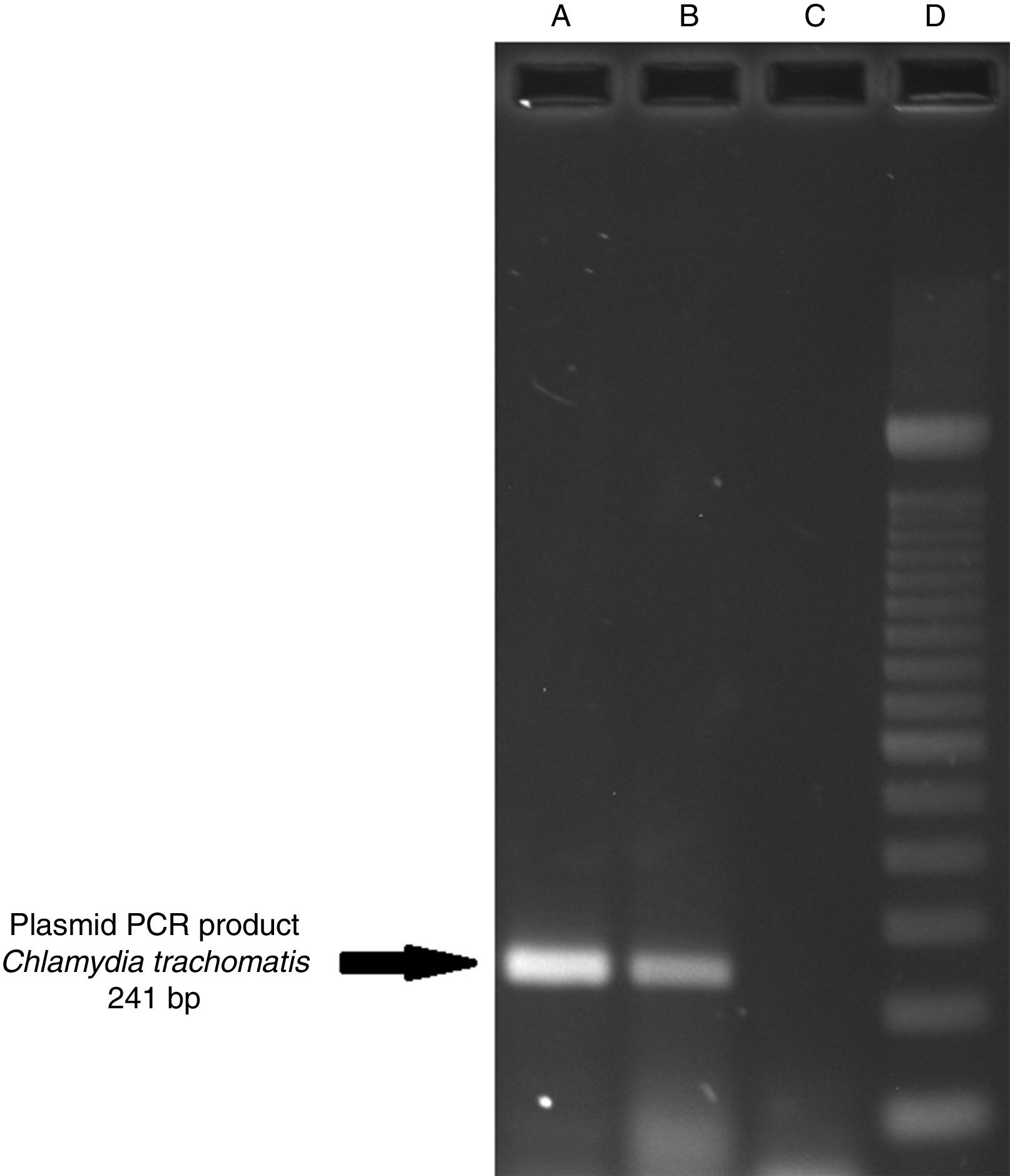
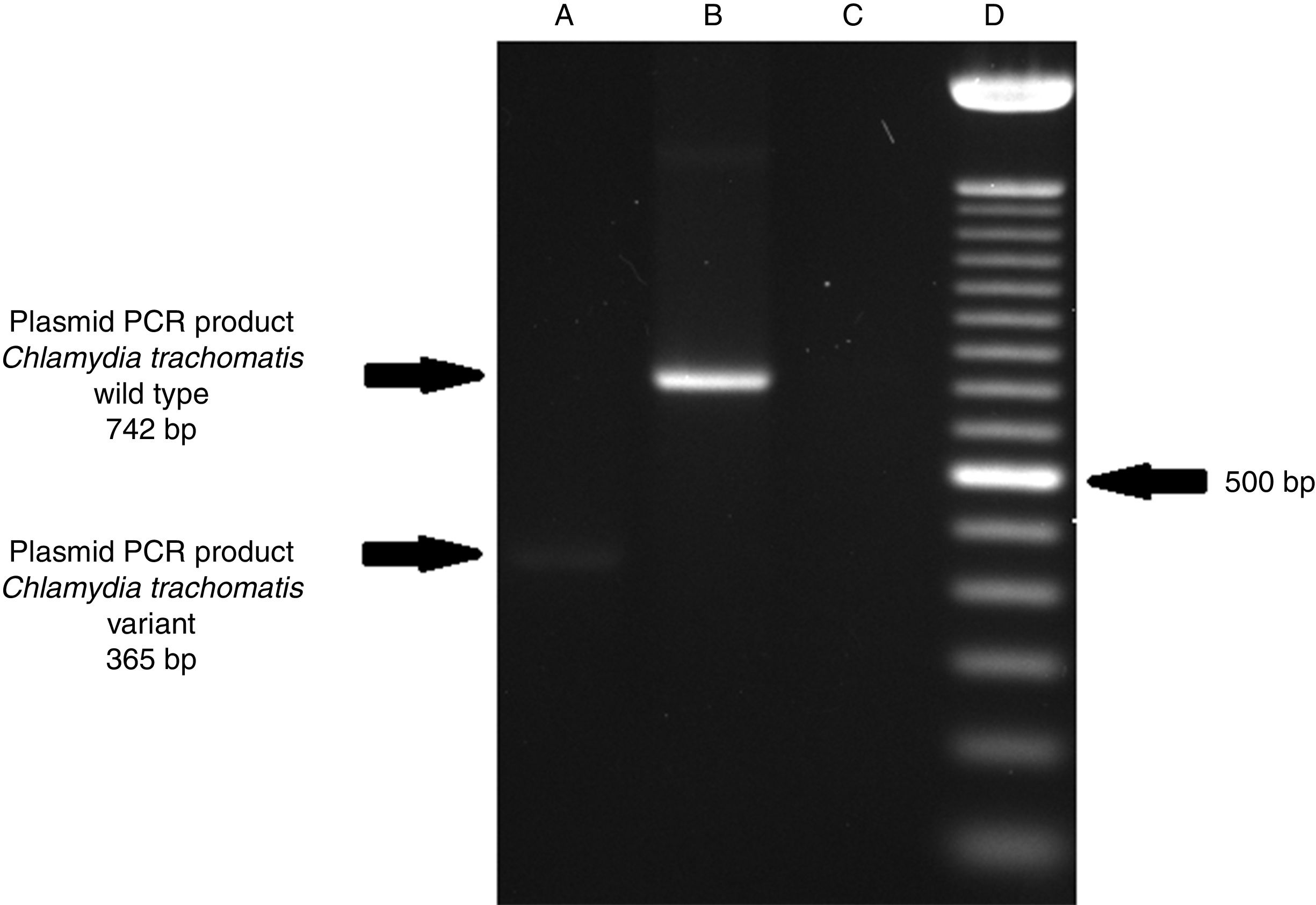
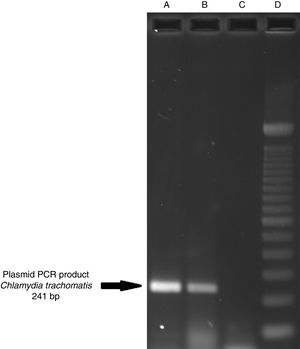
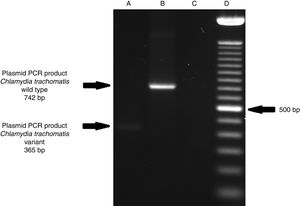
ResultsOf the 2339 endocervical samples analyzed by COBAS® Amplicor/Taqman48 assay, 69 were found to be positive for C. trachomatis, corresponding to a prevalence of 2.94%. The presence of C. trachomatis and its cryptic plasmid were confirmed by PCR. The expected product of 241bp from the ORF2 of the plasmid from C. trachomatis was obtained (Fig. 1). Of the 69 endocervical samples tested, the presence of the plasmid was confirmed in only 60 (88.23%). Deletion in ORF1 was investigated in the 60 clinical samples positive for C. trachomatis, and only one of these samples (m20) as a possible Swedish nvCT. In this case, a PCR product of 365bp was detected in sample m20 indicating the presence of the 377bp deletion, as shown in Fig. 2.
Detection of the cryptic plasmids of C. trachomatis in an endocervical sample. The image shows: Well (A) amplification with product of 241bp of the positive control (plasmid DNA from C. trachomatis ATCC® VR-902B), Well (B) PCR product of the clinical sample, Well (C) negative control (HeLa cell DNA), and Well (D) a 100bp DNA ladder (Invitrogen, CA, USA).
Detection of the Swedish nvCT in clinical samples by PCR. The image shows the PCR results used to determine the presence of 377bp deletions in the C. trachomatis cryptic plasmid. Well (A), indicates the presence of a 365bp product, which is associated with the reported 377bp deletion.2 Well (B), shows the presence of a 742bp product obtained from C. trachomatis with no plasmid deletion (plasmid DNA from C. trachomatis ATCC® VR-902B), Well (C), is the negative control (HeLa cell DNA), and Well (D) a 100bp DNA ladder (Invitrogen, CA, USA).
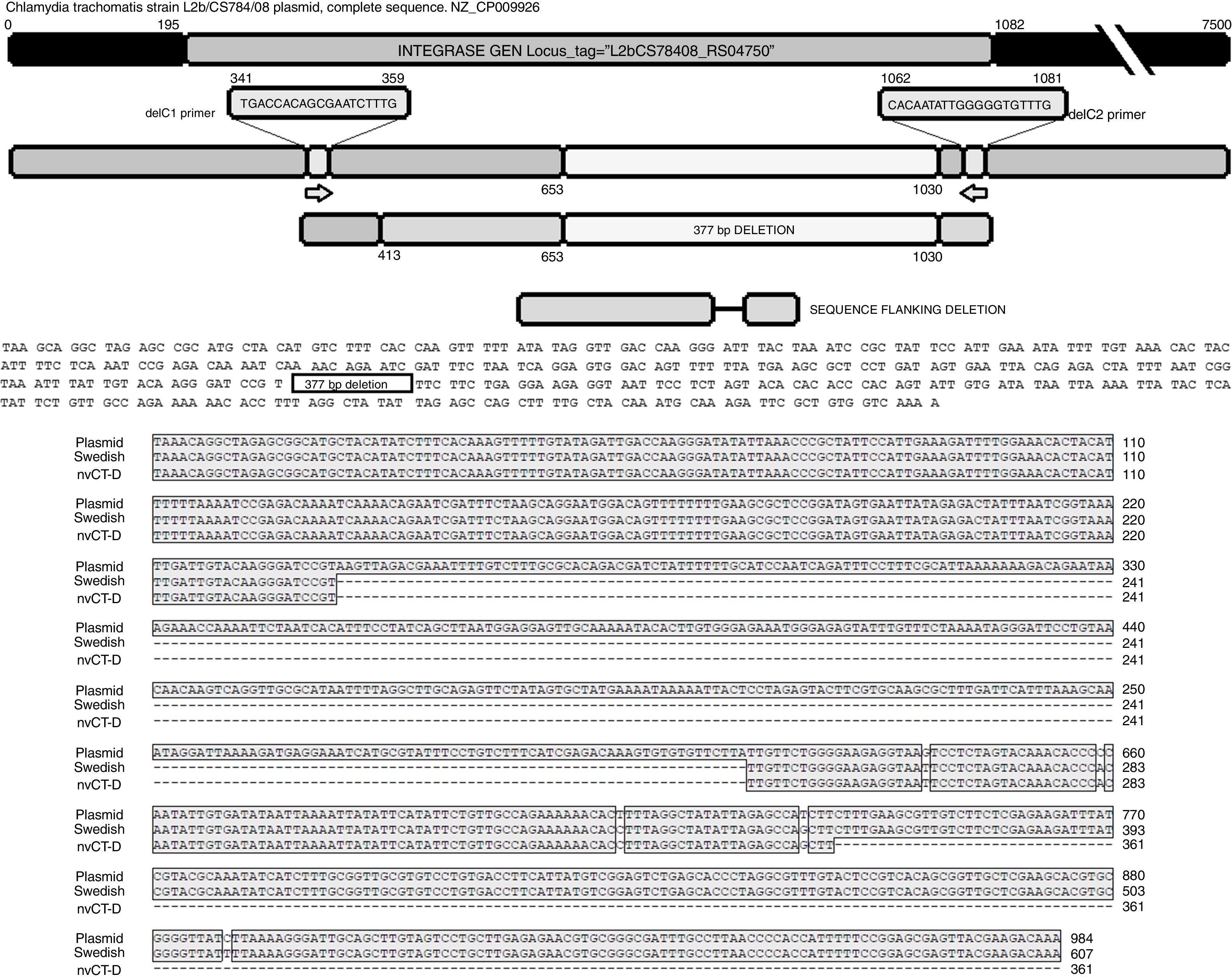
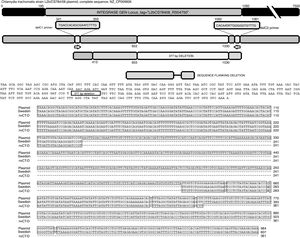
The sequence products (GenBank accession number: KY474387 and KY474388) obtained from possible Swedish-nvCT were analyzed with Blastn (version 2.3.1). Notably, a GenBank BLAST search indicated that the sequence of the clinical sample was like that of the plasmid of C. trachomatis in the flanking region where the deletion was reported, which demonstrated that the genetic motif of the lost 377bp spanned from nucleotide 653 to nucleotide 1030 of the plasmid (C. trachomatis strain L2b/CS784/08 plasmid, NZ_CP009926.1). This deletion was found in ORF1, which corresponds to an integrase (locus tag: L2bCS78408_RS04750) (Fig. 3). To confirm that this plasmid is of a Swedish nvCT, we amplified and analyzed a ORF3 region that show a 44bp perfect tandem duplication. The analysis of the ORF3 showed that in the variant found does not have the duplication of 44bp as the Swedish nvCT (Fig. 4).
Identification of the deletion site in the plasmid of C. trachomatis. The image shows the sequence of deletion site reported in GenBank (Swedish nvCT, GenBank: EF121757), the complete sequence of the plasmid (GenBank: NZ_CP009926), the sequence obtained in this study (nvCT-D; GenBank: KX300216, KY474387, and KY474388), and the hybridization sites of the primers (delc1 and delC2). Alignment revealed the previously reported deletion site, which involves the loss of 377bp (from nucleotides 653 to 1030) in the gene of the putative integrase in the C. trachomatis plasmid.
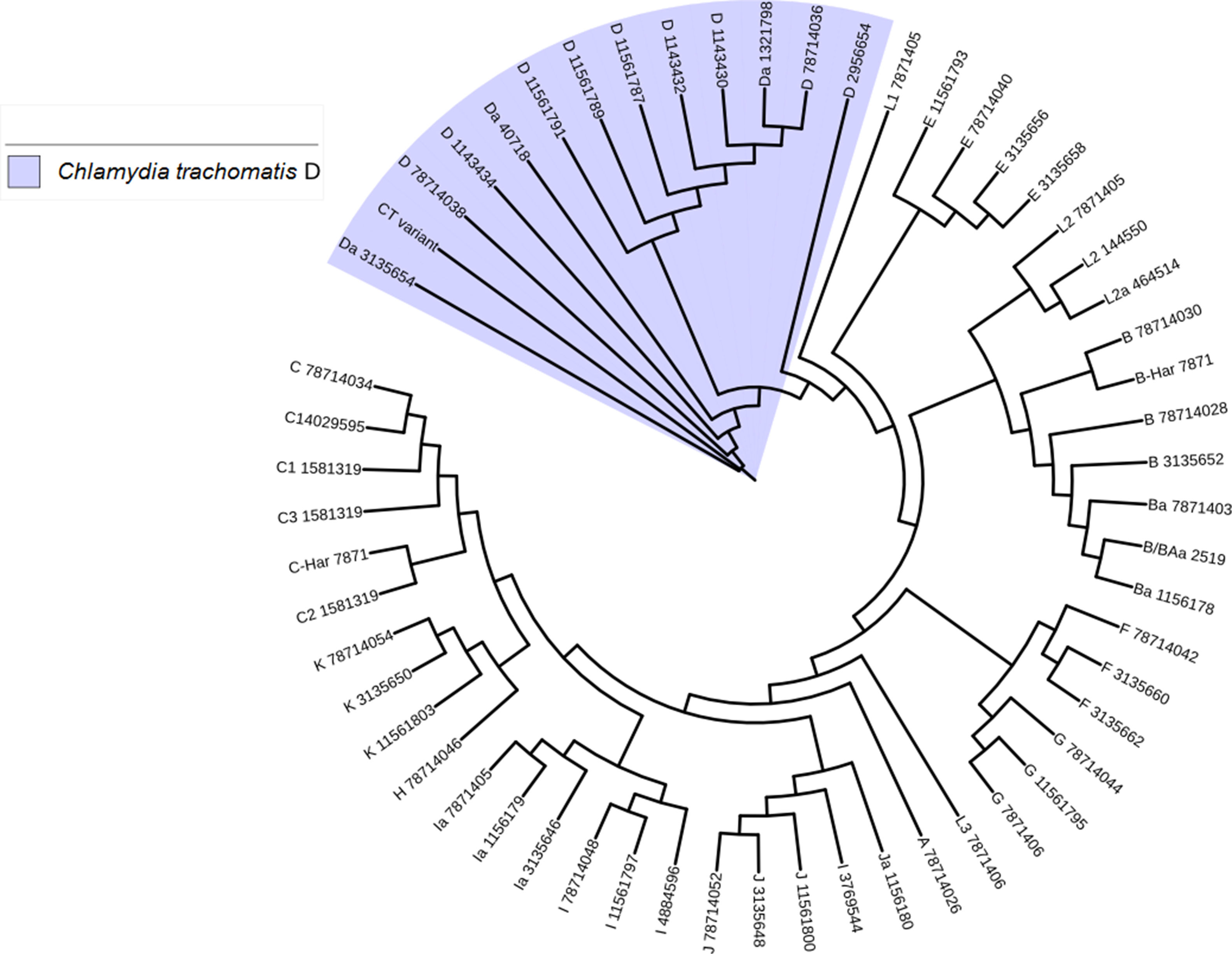
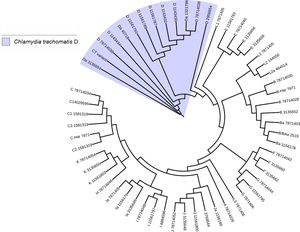
Another important aspect of this investigation was the identification of the C. trachomatis serovars of the samples that presented the deletion. This study was based specifically on the sequencing of the ompA gene. The sequence obtained (GenBank accession number: KY474386) was then aligned with the Genbank ompA sequences of all Chlamydia serovars reported to date. The phylogenetic tree based on ompA gene sequences indicated that the isolated obtained in this study belongs to serovar D of C. trachomatis, with a homology of 98% between sequences. (Fig. 5). To confirm this result, we amplified and analyzed the pmpH gene. The alignment results showed that this Mexican variant is a serotype D, and this localized into of the genito-urinary C. trachomatis pathotype (Fig. 6). The analysis of the chromosomal genes from ompA and pmpH, and of the ORF3 of the cryptic plasmid of both the Swedish nvCT and Mexican variant showed that are different. So it is presumed that this is a new variant of C. trachomatis (Mexican nvCT) of serotype D.
Phylogenetic tree of the sequences reported in GenBank for the ompA gene. The image shows the phylogenetic tree developed with iTOL. The sequences reported in GenBank and the Mexican nvCT identified in our study were analyzed with this program. The nucleotide sequence of the Mexican nvCT showed high homology with the cluster generated for serovar D of C. trachomatis.
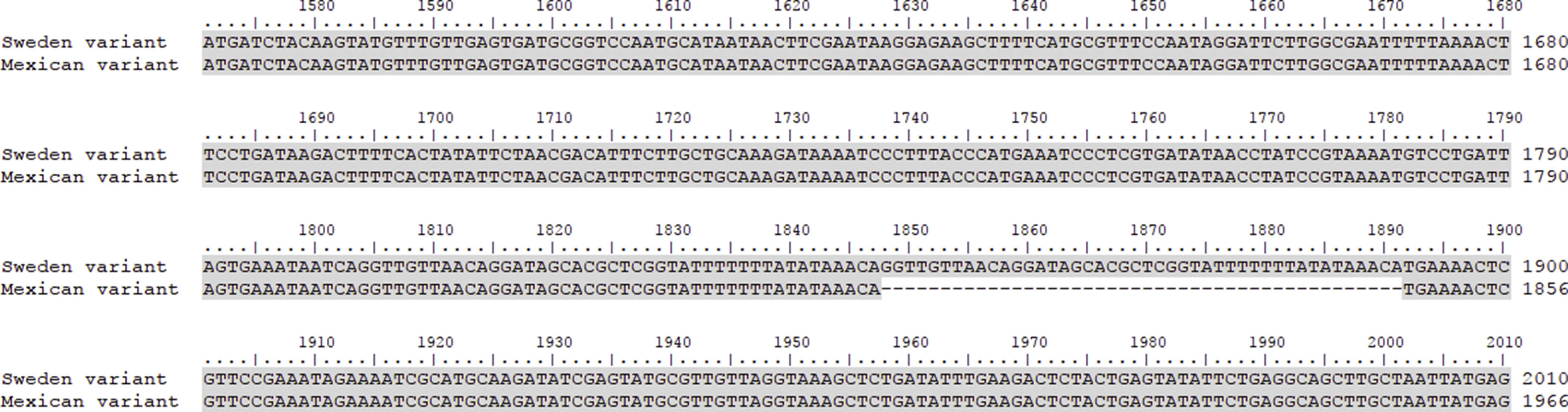
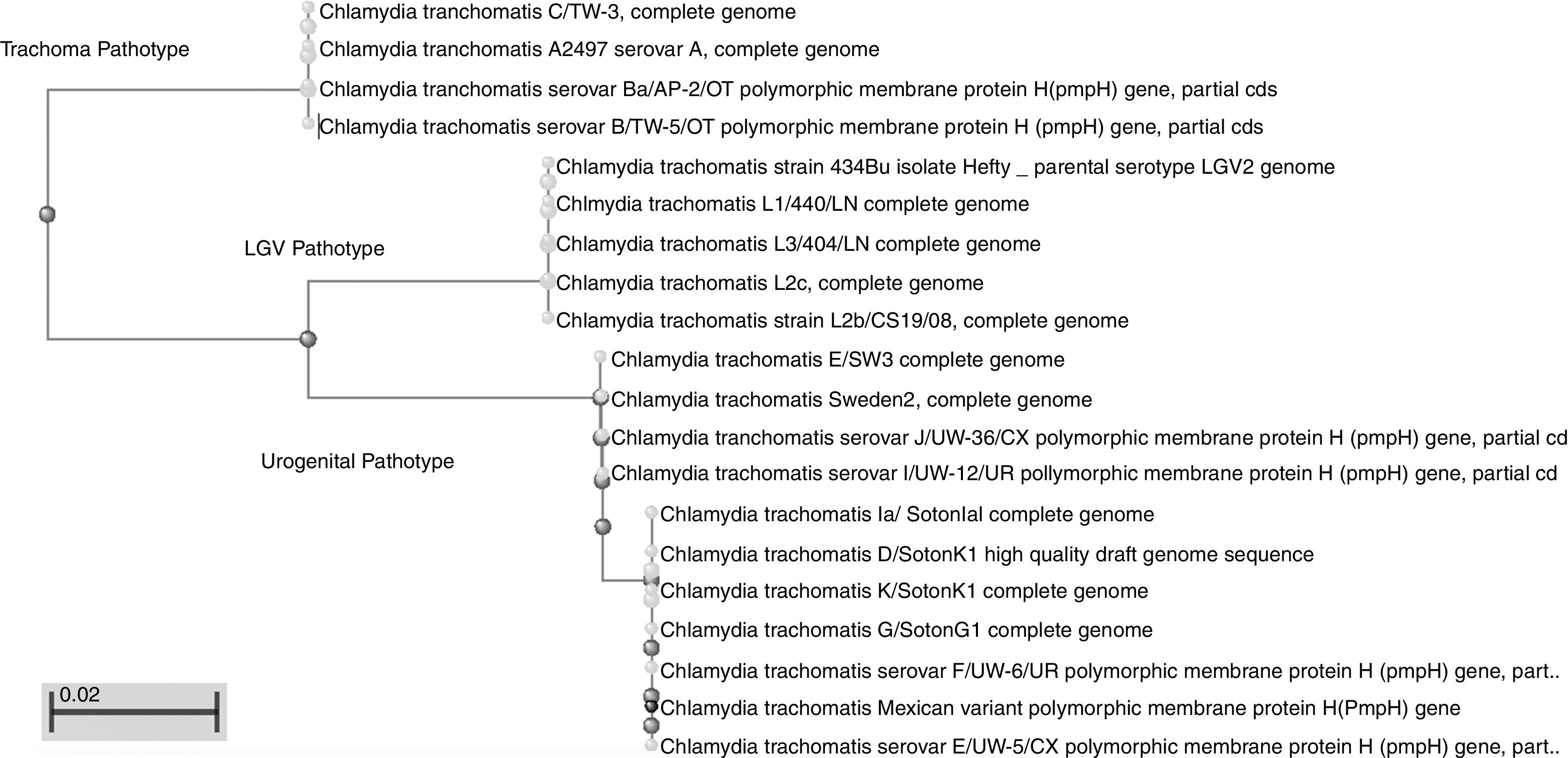
This work revealed several important aspects about the type of infection and the variants of C. trachomatis that develop them. First, an isolated with the same characteristics as the Swedish-nvCT was found. However, the region that shows a 44bp perfect tandem duplication into ORF3 of the cryptic plasmid is not present. Which assumes that the C. trachomatis found is a new variant. In addition, by phylogenetic analysis of the ompA gene (GenBank accession number: KY474386). These findings are supported by the results obtained by other investigations involving the genotyping of the ompA gene.7,13 This discovery is important because the Swedish nvCT belongs to serotype E.2,6 Likewise, the analysis of the pmpH gene grouped us to Mexican nvCT in the cluster of the C. trachomatis urogenital pathotype. Although Swedish nvCT belongs to the same pathotype that Mexican nvCT have significant differences that associate them with serovars E and D respectively (Fig. 7). These results confirm that the Mexican nvCT is a new variant.
The pmpH gene sequences of the Swedish nvCT, Mexican nvCT, C. trachomatis serovar E and C. trachomatis serovar D are shown. It is observed that Swedish nvCT and Mexican nvCT present differences in the sequence of the pmpH gene. In addition, these differences are associated with serovar E to nvCT-Swedish and serovar D to Mexican nvCT.
The prevalence determined for the Mexican nvCT in this study was 1.66% (i.e., 60C. trachomatis-positive samples from among 2339 samples tested). The number of positive samples for C. trachomatis obtains in this study (2.94%) is below the average of the reports.1,3 This information is important since several of the patients were previously treated against C. trachomatis in their clinics. Likewise, only the presence of the plasmid was determined in 61 of 69 (88.40%). The above was validated using the delC, KL, and ctorf3 primers (that hybridize with ORF 1, 2 and 3, respectively), where no amplification products were observed that confirmed the presence of the C. trachomatis cryptic plasmid. This suggest the presence of C. trachomatis strains plasmid-free that infected to Mexican population is in a major proportion that reported by Yeow et al.14 in women from Malaysia and by An et al. in Massachusetts, USA.14,15
An emerging theme is in vivo infections with plasmid-free C. trachomatis in asymptomatic patients or with a reduced pathology.16 Almost all clinical isolates of C. trachomatis contain a plasmid co-evolved with its genome; where this plasmid may play important roles in chlamydial pathogenesis.17 The high percentage obtained from C. trachomatis plasmid-free samples in this study could be due to a selective pressure by use of inadequate antimicrobial treatment. In 2007 reporting of a persistent chlamydia infection in a man that received azithromycin treatment.1 The molecular assays showed that the infection was by a plasmid-free C. trachomatis variant.1 The use of inadequate antimicrobial drugs during an infection is very common in Mexico. On several occasions without identification of infectious etiology, the patient takes an antimicrobial drug. Another possibility is the use of plasmid-based NAATs for systematic screening over a long period may result in diagnostic selection pressure and consequently the emergence of plasmid-free strains and false negative assay results. Some authors explain that the identification of plasmid-free C. trachomatis can be due to two possible conditions. That a plasmid-free strain has been identified or that the plasmid has been lost during the chronic infection process.15 Due to the above, is needed to carry out an investigation about the prevalence of plasmid-free C. trachomatis variant infection and detected the causes for elevated percentage in Mexico.
The alignment data obtained by BLASTn (NCBI) was used to identify the Mexican nvCT flanks 377-bp deletion. This genetic motif is in ORF1 of the plasmid, which is a putative integrase based on comparative genomics and proteomics analysis, but its role remains unknown in pathogenesis.2,18 Our studies revealed no difference between the sequences obtained (GenBank accession numbers: KY474387, KY474388, and KX300216) in our work and those reported for the Swedish nvCT (Fig. 3). Analyzing the sequence demonstrated 98.62% homology with the nucleotide sequences reported for the plasmid of C. trachomatis and 89.1% homology with respect to its amino acid sequence.
Serovar D is one of the more common serovars of C. trachomatis in terms of its worldwide prevalence.19,20 However, one difference between serovar E and serovar D is that the latter can produce a cytotoxin that provokes the rounding of infected cells because of the depolymerization of actin, and in a whole-organ fallopian tube model, the destruction of the epithelium in infected fallopian tubes has been observed.21,22 This suggests that in the evolution of the serovars of C. trachomatis, recombination events can occur, as occurred for genovar L2c, which is considered a recombinant strain of L2 and D serovars and has the functional toxin gene.23 Likewise, the study of comparative genomic analysis of 52 geographically diverse strains of C. trachomatis shows an extensive genome-wide recombination of this intracellular bacterial pathogen. With these data, we suggest that Mexican nvCT underwent a plasmid genetic recombination or plasmid exchange of the Swedish-nvCT to the Mexican nvCT.
In several European regions and the USA, the prevalence of non-invasive urogenital genotypes has been reported in patients with lymphogranuloma venereum (LGV).24–27 This could favor the selection of C. trachomatis variants and modify the local epidemiology. Therefore, the presence new variants of C. trachomatis must be searched for to determine the epidemiology of infection with this pathogen in Latin America.
Currently, no reports of the Swedish-nvCT in Mexico have been made. The only available studies have investigated the incidence and phylogeny of clinical isolates.5,7 The high migration rate of persons of European origin to Latin American countries and vice versa attributable to activities such as business, tourism, work, and studies represents a potential risk for the transmission of infectious diseases. Although the precise manner in which migration contributes to the spread of sexually transmitted infections is complex and poorly understood and increase the possibility of discovering new genovariants of C. trachomatis. Very small numbers of cases of infection with the Swedish-nvCT have been reported in European countries. For example, in Spain, only 0.4% of samples analyzed were positive for the new Swedish-nvCT, and similar values were obtained in Germany, France, and Russia.28,29 Higher prevalence rates have been reported in Nordic countries, whereas in the USA, the presence of the new Swedish-nvCT has not been reported.8,27 This is the first report of a genovars D of C. trachomatis with a deletion in the plasmid in the world that is likely to be the Swedish-nvCT.
Conflict of interestAll authors have seen and approved the manuscript being submitted. We confirm that the article is the authors’ original work. On behalf of all co-authors, the corresponding author shall bear responsibility for the work. We have no conflicts of interest to disclose.
The authors thank the Biology Institute (UNAM, CDMX, Mexico) for their support in the development of sequencing. This work was conducted in the National Institute of Perinatology and received no economic support.