Mycobacteria are a large group of microorganisms, multiple species of which are major causes of morbidity and mortality, such as tuberculosis and leprosy. At present, the emergence and spread of multidrug-resistant strains of Mycobacterium tuberculosis complex are one of the most serious health problems worldwide. Furthermore, in contrast to M. tuberculosis and Mycobacterium leprae, non-tuberculous mycobacteria (NTM) are more frequently isolated and, in many cases, treatment is based on drug susceptibility testing. This article is a review of the different methods to determine the in vitro drug susceptibility of M. tuberculosis complex and the most relevant NTM isolates. The molecular techniques currently used for rapid detection of resistance of clinical specimens are also analysed.
Las micobacterias son un amplio grupo de microorganismos en el que múltiples especies son causa de una importante morbimortalidad, como la tuberculosis y la lepra. La aparición y diseminación de cepas del complejo Mycobacterium tuberculosis resistentes a diversos fármacos constituye en la actualidad uno de los problemas sanitarios de mayor gravedad a nivel mundial. Por otro lado, las micobacterias diferentes de M. tuberculosis y Mycobacterium leprae, denominadas micobacterias no tuberculosas (MNT), son aislamientos cada vez más frecuentes, requiriendo en muchos casos un tratamiento que precisa una orientación sobre la sensibilidad de estos microorganismos a los antimicrobianos. En el presente artículo se revisan los métodos para determinar la sensibilidad in vitro a los antimicobacterianos de los aislamientos del complejo M. tuberculosis y las MNT más relevantes. Además, también se realiza un análisis de las técnicas moleculares de detección rápida de la resistencia a partir de las muestras clínicas.
Currently, more than 170 species of mycobacteria have been reported, and many of them are a major cause of morbidity and mortality in humans. Notable among them is tuberculosis (TB), caused by the Mycobacterium tuberculosis1 complex. According to the WHO, around 9.6 million people became ill from TB in 2014 and 1.5 million died from it, with it being the primary cause of death by an infectious agent.1 Also, in recent years, strains resistant to multiple drugs have appeared and are spreading. Thus, it is estimated that 480,000 people developed multidrug-resistant TB (MDR-TB; resistance to at least isoniazid and rifampicin) worldwide in 2014, of which 9% would have extended resistance (XDR-TB) to at least one of the second-line injectable drugs (capreomycin, kanamycin, or amikacin) and to a fluoroquinolone. Therefore, the rapid detection of resistance to antituberculosis drugs is fundamental to achieve adequate treatment and prevent the onset and spread of MDR-TB. Furthermore, nontuberculous mycobacteria (NTM) or environmental mycobacteria have become increasingly prevalent, with many of them being pathogenic (mycobacteriosis) and requiring a specific treatment that, in many cases, should be oriented by in vitro antimicrobial susceptibility testing.2–4
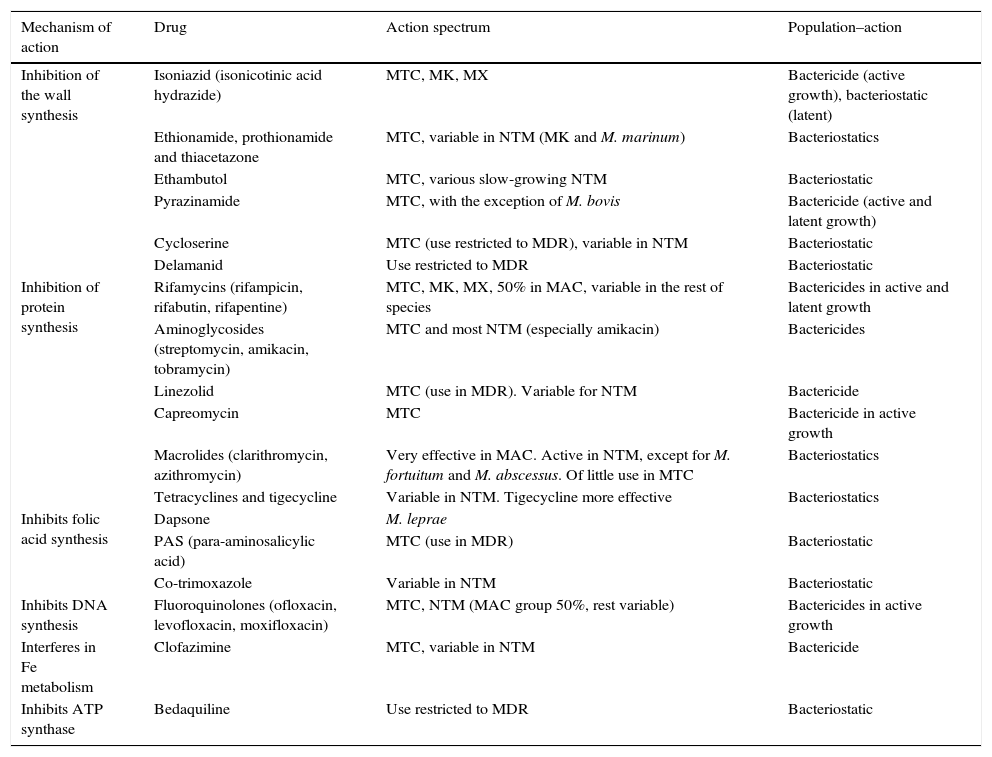
AntimicrobialsAlthough the therapeutic arsenal for mycobacterial infections is very limited, some new drugs are currently available, although not all have the same activity or are used to treat the same types of infection or different species (Table 1).5
Mechanism and action spectrum of antimycobacterials.
| Mechanism of action | Drug | Action spectrum | Population–action |
|---|---|---|---|
| Inhibition of the wall synthesis | Isoniazid (isonicotinic acid hydrazide) | MTC, MK, MX | Bactericide (active growth), bacteriostatic (latent) |
| Ethionamide, prothionamide and thiacetazone | MTC, variable in NTM (MK and M. marinum) | Bacteriostatics | |
| Ethambutol | MTC, various slow-growing NTM | Bacteriostatic | |
| Pyrazinamide | MTC, with the exception of M. bovis | Bactericide (active and latent growth) | |
| Cycloserine | MTC (use restricted to MDR), variable in NTM | Bacteriostatic | |
| Delamanid | Use restricted to MDR | Bacteriostatic | |
| Inhibition of protein synthesis | Rifamycins (rifampicin, rifabutin, rifapentine) | MTC, MK, MX, 50% in MAC, variable in the rest of species | Bactericides in active and latent growth |
| Aminoglycosides (streptomycin, amikacin, tobramycin) | MTC and most NTM (especially amikacin) | Bactericides | |
| Linezolid | MTC (use in MDR). Variable for NTM | Bactericide | |
| Capreomycin | MTC | Bactericide in active growth | |
| Macrolides (clarithromycin, azithromycin) | Very effective in MAC. Active in NTM, except for M. fortuitum and M. abscessus. Of little use in MTC | Bacteriostatics | |
| Tetracyclines and tigecycline | Variable in NTM. Tigecycline more effective | Bacteriostatics | |
| Inhibits folic acid synthesis | Dapsone | M. leprae | |
| PAS (para-aminosalicylic acid) | MTC (use in MDR) | Bacteriostatic | |
| Co-trimoxazole | Variable in NTM | Bacteriostatic | |
| Inhibits DNA synthesis | Fluoroquinolones (ofloxacin, levofloxacin, moxifloxacin) | MTC, NTM (MAC group 50%, rest variable) | Bactericides in active growth |
| Interferes in Fe metabolism | Clofazimine | MTC, variable in NTM | Bactericide |
| Inhibits ATP synthase | Bedaquiline | Use restricted to MDR | Bacteriostatic |
MAC, M. avium complex; MDR, multidrug-resistant tuberculosis; MK, M. kansasii; NTM, nontuberculous mycobacteria; MTC, M. tuberculosis complex; MX, M. xenopi.
Drugs called “first line” treatments include those administered as the first choice and are isoniazid (H), rifampicin (R), ethambutol (E), pyrazinamide (Z), and, for historical reasons, also streptomycin (S).5–7 The first four are those recommended by the WHO for a 6-month regimen (2HREZ+4HR). In cases of resistance, allergy, intolerance, hepatic toxicity or interaction with one or more of these drugs, second-line drugs should be used, including rifamycins (rifabutin, rifapentine), quinolones (ofloxacin, levofloxacin, moxifloxacin), aminoglycosides (amikacin, tobramycin), capreomycin, cycloserine, linezolid, isoniazid analogues (ethionamide, prothionamide), clofazimine, PAS or thiacetazone.5–7 In cases of MDR-TB or XDR-TB, recently developed drugs may also be used, such as bedaquiline and/or delamanid.8
Drugs used against nontuberculous mycobacteriaThe treatment of NTM, with the exception of leprosy, is less standardised than that of TB, with wide variations between species.2,9 For most slow-growing species, the mainstays of treatment are clarithromycin, ethambutol and amikacin, complemented with other drugs such as quinolones, rifamycins, linezolid, and probably, with a lesser degree of efficacy, cephalosporins, carbapenems and tetracyclines. Mycobacterium kansasii and Mycobacterium xenopi infections are treated similarly to TB, except for pyrazinamide, since it does not have activity against NTM.2,9 Rapidly growing species are resistant to classic antituberculosis drugs and also have clarithromycin as the mainstay of treatment, with the exception of Mycobacterium fortuitum, which is resistant, as well as 80–85% of Mycobacterium abscessus isolates.2,9
Drug susceptibility testing in M. tuberculosis complexThe history of susceptibility testing for the M. tuberculosis complex arose parallel to the development of specific pharmacological treatment for TB, which began with the discovery of streptomycin in 1944. From the beginning, it was observed that monotherapy achieved an initial clinical improvement followed by a relapse of the disease. This phenomenon, known as ‘fall and rise’, was due to the selection of resistant bacteria. In 2000, the WHO defined resistance in various categories from an epidemiological point of view: (a) Resistance in new cases: strains isolated in patients who have never received antituberculosis treatment for more than one month. (b) Resistance in treated cases: strains isolated in patients who have previously received antituberculosis treatment for more than one month. (c) Multidrug resistance (MDR-TB): joint resistance to at least isoniazid and rifampicin. (d) Poly-resistance: resistance to more than one drug not including isoniazid and rifampicin at the same time. (e) Combined resistance: sum of all resistance types in a specific area. This indicates the resistant strain load in a community. In 2006, the category of Extended resistance (XDR-TB) was added, which was mentioned in the introduction.
The definitive confirmation of M. tuberculosis complex drug resistance must always be conducted using microbiological methods. In studies in the 1960s by Canetti et al.,10 the frequency of spontaneous mutations associated with each drug was established. In the case of isoniazid, ethambutol and streptomycin, a resistant mutant appeared in one out of 10−5–10−7 bacteria, for rifampicin in one out of 10−7–10−9 and for pyrazinamide in one out of 10−2–10−4. The theoretical frequency of mutants resistant to more than one drug would be the exponential sum of the individual mutants resistant to each drug. The bacterial populations present in the lesions were also estimated. Thus, small infiltrates had populations of 103–105 bacteria, and cavities could reach 107–109 bacteria which makes it very improbable that simultaneous mutations to more than one drug would occur spontaneously. TB is deemed resistant when 1% or more of the bacterial population is resistant to a certain drug.10
The critical concentrations of the different drugs were established by international consensus and represent the lowest concentrations that inhibit the growth of “wild strains” (susceptible strains) of the M. tuberculosis complex that have never been exposed to said drugs, while not inhibiting the strains from patients that do not respond to treatment, considered resistant. They were initially postulated for the Löwenstein–Jensen medium, and later equivalencies were established for Middlebrook 7H10 and 7H11 agar media, as well as for automated liquid media culture systems.
In vitro susceptibility testing in M. tuberculosis complex must be performed on the first isolate obtained from a new patient, in cases of suspected therapeutic failure and before the disease relapses.7,11–13 Also, when the TB simultaneously affects more than one organ, the tests must be done individually for each location, without assuming that the results will always coincide. All first-line drugs should be included (isoniazid, rifampicin, pyrazinamide, ethambutol and streptomycin), since they provide information about the therapeutic scheme currently recommended for most patients.7,11–14 Traditional second-line drugs (amikacin, capreomycin, clofazimine, cycloserine, ethionamide, kanamycin and PAS) and new ones, such as fluoroquinolones (ciprofloxacin, levofloxacin, moxifloxacin), linezolid, and, in certain cases, rifabutin, must be evaluated whenever: (a) a strain is resistant to isoniazid, rifampicin, ethambutol or pyrazinamide; (b) when mutations in genes that encode resistance to isoniazid and/or rifampicin are detected using molecular techniques; and (c) when clinical-epidemiological information is available that recommends doing so.13–15
Solid culture media-based methodsThe absolute concentrations method, described by Meissner, is based on the comparison of the number of colonies in the presence of the drug to the number of colonies in the culture medium without the drug. The results are affected by the size of the inoculum and by the precision in the definition of the critical concentration of each drug. The resistance ratio method, described by Mitchison, compares the minimum inhibitory concentration (MIC) of a specific strain to that of a reference strain. The results depend on the adequate standardisation of the drugs and the inoculum, although the critical concentrations do not need to be defined. The multiple proportion method, described by Canetti et al.10 in 1963, shows the proportion of resistant bacilli present in a culture. Micro-organism resistance is considered clinically significant when the resistant bacterial population exceeds 1%. This is the most precise method and the one that produces fewest false results, which is why it is the most widely used worldwide.7,11,13,16 The epsilometer (E-test) method is based on a gradient of antimicrobial diffusion combining the principles of disc diffusion and agar dilution in the study of in vitro susceptibility. It is a quantitative technique to determine the MIC of antimicrobials.17
Liquid culture media-based methodsThere are currently two nonradiometric systems for susceptibility testing: BACTEC™ MGIT™ 960 (Becton-Dickinson Diagnostics, Sparks, MD, USA) and VersaTREK® (Thermo Scientific, Westlake, OH, USA).7,11,13,18 Both use supplemented Middlebrook 7H9 medium, and the growth is shown via detection of the consumption/production of certain gases. The initial and secondary critical concentrations that are analysed come predetermined by the manufacturer for first-line drugs. Although the detection of MDR-TB does not usually pose any problems, there are some inter-laboratory discrepancies with streptomycin, ethambutol and pyrazinamide. In the case of pyrazinamide, since it is active in acidic media (pH 5.5), the pH of the culture medium must match these conditions, despite the fact that approximately 10% of M. tuberculosis isolates do not grow well at this pH. On the other hand, it is also possible to analyse critical concentrations of other second-line drugs if the assessed substance is available.
Alternative susceptibility methodsIn mycobacteriophage-based techniques, when adding the phages to a culture in the presence of an antibiotic, the phages will only infect the viable mycobacteria, which are the resistant ones. These have been applied in isoniazid and rifampicin susceptibility tests, but they present problems with susceptibility, specificity and a high rate of contamination. Flow cytometry, using cell permeable nucleic acid stains, shows viable mycobacteria (the control mycobacteria and the drug-resistant ones) with higher levels of fluorescence compared to the non-viable ones (drug-susceptible). One of the drawbacks is the high cost of the equipment. Broth microdilution (Mycotb Sensititre® Thermo Scientific) allows for the MIC of multiple drugs to be determined. The greatest difficulty lies in reading and interpreting the results, since the growth of these bacteria is not homogeneous. The nitrate reductase test is based on the capacity of the M. tuberculosis complex to reduce nitrates to nitrites. Tubes of Löwenstein–Jensen medium incorporating KNO3, with and without drugs, are used for this purpose. Microscopic-observation drug-susceptibility (MODS) is based on the early detection of the cord factor exhibited by the M. tuberculosis complex, with the help of an inverted microscope. Resazurin microtiter assay (REMA) is an indirect method based on the detection of colour changes in the culture medium due to oxidation (blue) and reduction (pink) of resazurin.
The WHO considers that nitrite reductase, MODS and REMA are simple, cheap techniques that can be used for rapid screening for resistance to isoniazid and rifampicin in countries with limited resources.11,13,19 Susceptibility and specificity data exceed 95%.
Antimicrobial susceptibility testing in nontuberculous mycobacteriaAlthough more than 160 species of NTM have been reported, susceptibility testing is only recommended in clinically-significant isolates for which a correlation has been shown between in vitro and in vivo results.2,13 Thus, the Clinical and Laboratory Standards Institute (CLSI) establishes recommendations only for the group comprising Mycobacterium avium complex, M. kansasii, Mycobacterium marinum and non-pigmented rapidly growing mycobacteria.7
Susceptibility testing in slow-growing nontuberculous mycobacteriaIn general, the methodology of choice that CLSI recommends is microdilution in cation-adjusted Mueller Hinton (CAMH) broth, supplemented with OADC or ADC. However, in some cases there are alternatives to this methodology.
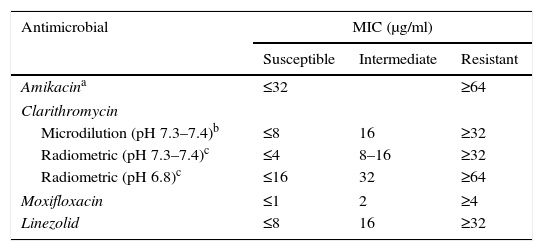
Recommendations for M. avium complexThe treatment of M. avium complex (MAC) is based on the use of clarithromycin as an essential or primary drug, and ethambutol and rifampicin (or rifabutin) as adjunct therapies, although there are data showing that other antibiotics (amikacin, linezolid and moxifloxacin) could be useful in some cases. Given that a clinical correlation has only been demonstrated with in vitro tests for macrolides, the CLSI recommends that only these drugs be tested.7 However, when resistance to clarithromycin is present, it is reasonable to test other drugs (moxifloxacin, linezolid and aminoglycosides). Clarithromycin's activity is very sensitive to the culture medium and its pH, therefore the MICs used generally as cut-off points correspond to a CAMH medium at a slightly basic pH of 7.3–7.4. Another method that can be used is macrodilution using the BACTEC™, MGIT™ 960 and VersaTREK® systems.2,13 There is currently a commercial microdilution technique in microtitration plates (Slomyco, Sensititre®, Thermo Scientific), that allows for simple, reproducible in vitro susceptibility testing with a standard incubation of 7 days. Although it includes 13 antimicrobials with multiple concentrations, it is important to maintain the previously stated interpretation criteria (Table 2).
Interpretation criteria for susceptibility testing of the M. avium complex via broth microdilution.
| Antimicrobial | MIC (μg/ml) | ||
|---|---|---|---|
| Susceptible | Intermediate | Resistant | |
| Amikacina | ≤32 | ≥64 | |
| Clarithromycin | |||
| Microdilution (pH 7.3–7.4)b | ≤8 | 16 | ≥32 |
| Radiometric (pH 7.3–7.4)c | ≤4 | 8–16 | ≥32 |
| Radiometric (pH 6.8)c | ≤16 | 32 | ≥64 |
| Moxifloxacin | ≤1 | 2 | ≥4 |
| Linezolid | ≤8 | 16 | ≥32 |
Standard treatments for infections caused by this mycobacteria include rifampicin (or rifabutin), isoniazid, and a third drug (ethambutol or streptomycin) for 18 months (at least 12 months of negative cultures). In cases of possible resistance, susceptibility should be conducted in vitro for these and other potentially-active antimicrobials (clarithromycin, moxifloxacin or levofloxacin, amikacin, linezolid and rifabutin).2,7,13 One aspect to highlight is that M. kansasii naturally presents a somewhat decreased susceptibility to isoniazid in comparison with M. tuberculosis, although it has clinical activity.
The recommended method is microdilution, although the proportion method in Middlebrook 7H10 agar and automated nonradiometric systems have been used. As mentioned previously in MAC, there is currently a commercial microdilution technique that, although it includes multiple antimicrobials with diverse concentrations, it is important to follow the current interpretation criteria (CLSI).
Recommendations for M. marinumAntimicrobial susceptibility testing is currently only recommended for this species when there is a clinical and microbiological failure, using the broth microdilution method at 28–30°C for 7 days.2,7,13
Other slow-growing nontuberculous mycobacteriaGenerally, it is recommended to perform in vitro susceptibility testing using broth microdilution with the same drugs and the same concentrations as for M. kansasii for all species with good growth (non-deficient) and in those in which therapeutic failure and persistent positive cultures are detected.
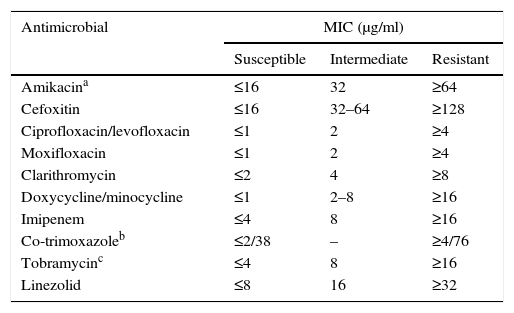
Susceptibility testing in rapid-growing nontuberculous mycobacteriaTreatment for these micro-organisms is usually conducted with drugs against which the remaining species are resistant or, at least, have a lesser susceptibility. Due to all of this, in vitro susceptibility tests have certain differentiating features from the rest of the mycobacteria. The recommended technique is broth microdilution with an incubation of 3–4 days at 30°C (Table 3).2,3,7,13 Other methods present interpretation problems and should not be used. We must keep in mind the possible presence of inducible erm methylases. Therefore, reincubating the plates for up to 14 days and assessing the increase of the macrolides’ MIC is recommended to detect this phenomenon. There is currently a commercial microdilution technique designed specifically for rapid-growing mycobacteria and aerobic actinomycetes (Rapmyco, Sensititre®, Thermo Scientific).
Interpretation criteria for susceptibility testing of rapid-growing mycobacteria via broth microdilution.
| Antimicrobial | MIC (μg/ml) | ||
|---|---|---|---|
| Susceptible | Intermediate | Resistant | |
| Amikacina | ≤16 | 32 | ≥64 |
| Cefoxitin | ≤16 | 32–64 | ≥128 |
| Ciprofloxacin/levofloxacin | ≤1 | 2 | ≥4 |
| Moxifloxacin | ≤1 | 2 | ≥4 |
| Clarithromycin | ≤2 | 4 | ≥8 |
| Doxycycline/minocycline | ≤1 | 2–8 | ≥16 |
| Imipenem | ≤4 | 8 | ≥16 |
| Co-trimoxazoleb | ≤2/38 | – | ≥4/76 |
| Tobramycinc | ≤4 | 8 | ≥16 |
| Linezolid | ≤8 | 16 | ≥32 |
Antibacterial resistance is mainly due to chromosomal mutations in genes that encode their target or the activating enzymes. They can be spontaneous or drug-induced after exposure to monotherapy.2,3,20,21
IsoniazidResistance is associated above all with mutations in 2 genes (68–90%): (a) the katG gene, that is responsible for activating the drug; the most common mutation affects codon 315 and is related to high-level resistance (60–70% of resistance), and (b) the inhA gene, which intervenes in the synthesis of the bacterial wall. Mutations (8–20%) cause low-level and cross-resistance with ethionamide.13,20,21 NTM, except for M. kansasii and M. xenopi are resistant due to incapacity of activation of the drug.2,3,13
Rifampicin and other rifamycinsResistance to the M. tuberculosis complex is related to mutations in the rpoB gene (>95%), involved in protein synthesis. Codons 526 and 531 are the most affected and confer high-level and cross-resistance with other rifamycins.13,20,21 Mutations in some codons will maintain susceptibility to rifabutin. Similar mutations in NTM have been described.
Aminoglycosides and capreomycinResistance is based on ribosomal mutations involved in protein synthesis. Resistance to streptomycin is associated with mutations (50–60%) in the rpsL and rrs genes (codons 530–912), while resistance to amikacin, kanamycin, and capreomycin is associated with mutations in the rrs gene (codons 1400–1500).13,20,21 There is partial cross-resistance between them, with isolated resistance to streptomycin that may be susceptible to the others. The tlyA gene has been linked to resistance to capreomycin and the eis gene has been linked to resistance to kanamycin.
EthambutolResistance is related to mutations in the embB gene, which is involved in bacterial wall synthesis. The majority (60–80%) have been described in codons 306 and 406.13,20,21 Some species of NTM have natural resistance.2,3
PyrazinamideResistance is associated with mutations throughout the pncA gene (72–97%) that regulates drug activation. Mycobacterium bovis and its BGC variant, as well as NTM, have natural resistance.2,3,13,20,21
FluoroquinolonesThe most frequent mechanism is mutations in the gyrA gene (85%) and the gyrB gene (10%). There is cross-resistance between fluoroquinolones, but not full resistance, depending on the affected codons.2,3,13,20,21
Macrolides and ketolidesResistance is linked to mutations in the 23S ribosomal gene and, above all, to inducible methylases regulated by the erm gene. These are observed in species such as M. fortuitum and M. abscessus subsp. abscessus and subsp. bolletii.2
Beta-lactamsResistance is associated with a decrease in the permeability and affinity of the PBPs and, mainly, to beta-lactamases.2,3
Tetracyclines and glycylglycinesResistance to tetracyclines is due to ribosomal protection proteins or efflux pumps.2,3 To date, resistance to tigecycline has not been demonstrated.
OxazolidinonesThe resistance of mycobacteria to linezolid is associated with mutations in the rRNA 23S gene.13,20
Co-trimoxazoleThe resistance mechanism is not clear in mycobacteria.
Bedaquiline and delamanidThe resistance to bedaquiline has been associated with a mutation in the mmpR gene and the resistance to delamanid has been associated with the fbiA and fgd1 genes.22
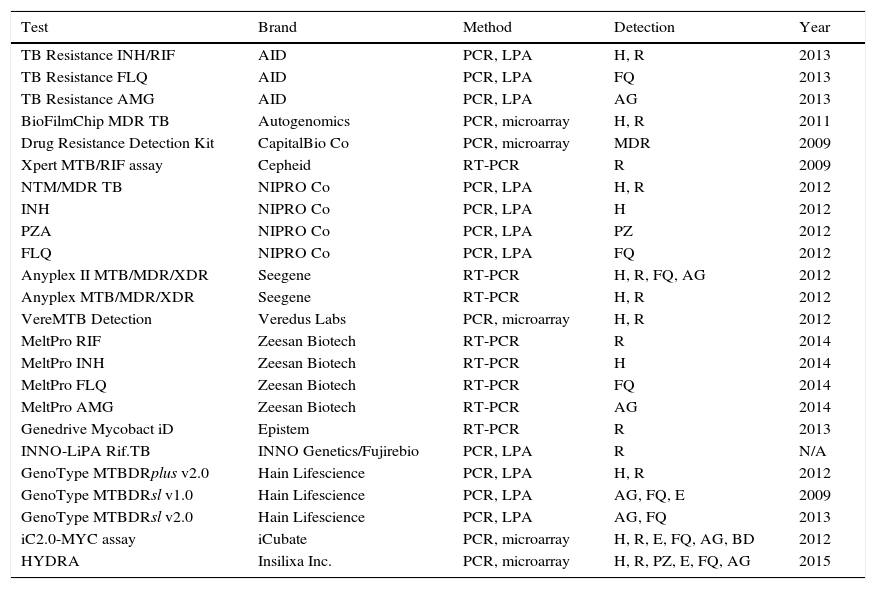
Molecular detection of resistancesThe reference method for determining resistance to antituberculosis drugs is the conventional phenotypic susceptibility test, although the results tend to take several weeks.7,11,13 Molecular detection of resistance, based on showing chromosomal mutations, offers quick and useful results.21,23 Commercial techniques are those which are most widely used due to their greater standardisation (Table 4). The most used are Xpert® MTB/RIF Assay (Cepheid, Sunnyvale, CA, USA) and GenoType® MTBDRplus (Hain Lifescience, Nehren, Germany), both approved by the WHO for the rapid diagnosis of MDR-TB. Xpert® MTB/RIF allows for diagnosis of the M. tuberculosis complex and the detection of mutations resistant to rifampicin in <2h, with minimal manipulation, limited to inserting the sample into a cartridge where the entire amplification and detection process is performed. The system uses five fluorescent probes covering the natural sequence of the 81pb region of the rpoB gene where more than 95% of resistances to rifampicin occur. For the diagnosis of pulmonary and extrapulmonary TB, susceptibility is 95–98% in samples with positive microscopy and 67–69% with negative microscopy, taking the culture as reference.24 The specificity is 99%. In synthesis, Xpert MTB/RIF® provides 23% more positive cases than microscopy. Its greatest drawbacks are that it only detects resistance to rifampicin and its financial cost, although for countries with limited resources, the cost is 6–7 times lower (less than €10).24 The GenoType® MTBDRplus is based on amplification and reverse hybridisation on nitrocellulose strips that contain immobilised probes corresponding to natural sequences and sequences of the most frequent mutations in the rpoB (rifampicin) and katG and inhA (isoniazid) genes. Currently, it can be used based on cultures or clinical samples, even with negative bacilloscopy. Various meta-analyses establish a global susceptibility of 98% for resistance to rifampicin and 84.3% for isoniazid, with 98–99.5% specificity. Samples with negative microscopy can have 5–12% invalid results due to low susceptibility. Results that are not interpretable due to the simultaneous absence of natural and mutated bands in the same locus, due to mutations not contained in the kit have been reported. Another possibility is a discrepant result with the sensitive genotype and resistant phenotype, due to mechanisms other than those detected by the commercial product. The resistant genotype and sensitive phenotype option would be caused by the coexistence of mixed or heteroresistant populations or low-level resistances, that have been described in 4–13% of cases.25 At least 50% of patients with MDR-TB are resistant to other drugs. Tests have also been marketed to detect mutations of resistance to second-line drugs. The most well-known product is GenoType® MTBDRsl (Hain Lifescience). Two versions exist: v1.0 and v2.0. The first is directed at mutations of resistance to capreomycin and the aminoglycosides amikacin and kanamycin in the rrs gene (codons 1401 and 1484), as well as to fluoroquinolones in the gyrA gene (codons 90, 91 and 94), and to ethambutol in the embB gene (codon 306). A recent analysis observed good susceptibility to fluoroquinolones (83–85%), but lower susceptibility to aminoglycosides (77%), attributable to kanamycin (69.9%).26 Version v2.0 seeks to improve yield including the gyrB gene for fluoroquinolones and the eis gene for kanamycin, suppressing the ethambutol of the kit. It has recently been evaluated with a susceptibility of 96% to kanamycin. However, a decrease in specificity for injectables (91%) has also been observed, due to the detection of mutations of still unknown significance in the eis gene in some susceptible strains.27 Aside from the above, the following products are also being distributed to a certain extent: INNO-Lipa® RifTB (INNO Genetics, Gent, Belgium/Fujirebio, Tokyo, Japan), Anyplex II™ MTB/MDR/XDR (Seegene, Seoul, Korea) and the TB Resistance® Assay (AID; Autoimmun Diagnostika, Strassberg, Germany).18,21,23 Although not discounting phenotypic susceptibility tests, the main use of molecular resistance detection methods, in our environment, is to quickly confirm or rule out, and to a greater or lesser extent, based on the drugs, resistance in cases of high suspicion. Recently, a method has been marketed (GenoType® NTM-DR, Hain Lifescience) that, in addition to identifying some NTM species, allows detection of the erm gene in M. abscessus, and of the mutations in the rrs (aminoglycosides) and/or rrl (macrolides) genes.
Marketed methods for the detection of resistant mutations to antituberculosis drugs.
| Test | Brand | Method | Detection | Year |
|---|---|---|---|---|
| TB Resistance INH/RIF | AID | PCR, LPA | H, R | 2013 |
| TB Resistance FLQ | AID | PCR, LPA | FQ | 2013 |
| TB Resistance AMG | AID | PCR, LPA | AG | 2013 |
| BioFilmChip MDR TB | Autogenomics | PCR, microarray | H, R | 2011 |
| Drug Resistance Detection Kit | CapitalBio Co | PCR, microarray | MDR | 2009 |
| Xpert MTB/RIF assay | Cepheid | RT-PCR | R | 2009 |
| NTM/MDR TB | NIPRO Co | PCR, LPA | H, R | 2012 |
| INH | NIPRO Co | PCR, LPA | H | 2012 |
| PZA | NIPRO Co | PCR, LPA | PZ | 2012 |
| FLQ | NIPRO Co | PCR, LPA | FQ | 2012 |
| Anyplex II MTB/MDR/XDR | Seegene | RT-PCR | H, R, FQ, AG | 2012 |
| Anyplex MTB/MDR/XDR | Seegene | RT-PCR | H, R | 2012 |
| VereMTB Detection | Veredus Labs | PCR, microarray | H, R | 2012 |
| MeltPro RIF | Zeesan Biotech | RT-PCR | R | 2014 |
| MeltPro INH | Zeesan Biotech | RT-PCR | H | 2014 |
| MeltPro FLQ | Zeesan Biotech | RT-PCR | FQ | 2014 |
| MeltPro AMG | Zeesan Biotech | RT-PCR | AG | 2014 |
| Genedrive Mycobact iD | Epistem | RT-PCR | R | 2013 |
| INNO-LiPA Rif.TB | INNO Genetics/Fujirebio | PCR, LPA | R | N/A |
| GenoType MTBDRplus v2.0 | Hain Lifescience | PCR, LPA | H, R | 2012 |
| GenoType MTBDRsl v1.0 | Hain Lifescience | PCR, LPA | AG, FQ, E | 2009 |
| GenoType MTBDRsl v2.0 | Hain Lifescience | PCR, LPA | AG, FQ | 2013 |
| iC2.0-MYC assay | iCubate | PCR, microarray | H, R, E, FQ, AG, BD | 2012 |
| HYDRA | Insilixa Inc. | PCR, microarray | H, R, PZ, E, FQ, AG | 2015 |
AG, aminoglycosides; BD, bedaquiline; E, ethambutol; FQ, fluoroquinolones; H, isoniazid; LPA, line probe assay; N/A, not available; PCR, polymerase chain reaction; PZ, pyrazinamide; R, rifampicin; RT-PCR, real-time PCR.
Modified from Tuberculosis Diagnostics Technology and Market Landscape, UNITAID WHO 2015.
The authors have declared that they have no conflicts of interest.
Please cite this article as: Alcaide F, Esteban J, González-Martin J, Palacios J-J. Métodos de determinación de sensibilidad a los antimicrobianos en micobacterias. Enferm Infecc Microbiol Clin. 2017;35:527–533.