Streptococcus agalactiae, or group B streptococci (GBS), is the main aetiological agent of early neonatal sepsis in developed countries. This microorganism belongs to the gastrointestinal tract microbiota wherefrom it can colonize the vagina and be vertically transmitted to the child either before or at birth, and subsequently cause infection in the newborn. Approximately, 50% of newborns born to women with GBS become colonized, with 1–2% developing early neonatal infection if no preventive intervention is performed. The aim of this study was to characterize and compare serotypes, virulence factors and antimicrobial resistance of GBS isolates collected from pregnant women and newborns in several hospitals in Catalonia.
Methods242 GBS strains were analyzed including 95 colonizers and 68 pathogenic strains isolated from pregnant women, and 79 strains isolated from neonates with sepsis in order to determine serotype, virulence and antimicrobial resistance.
ResultsSerotype distribution was different among the three groups, with serotypes Ia and II being significantly more frequent among colonizing strains (p=0.001 and 0.012, respectively). Virulence factors bca and scpB were significantly more frequent among neonatal strains than pathogenic or colonizing strains (p=0.0001 and 0.002, respectively). Pathogenic strains were significantly more resistant to erythromycin, clindamycin and azithromycin than their non-pathogenic counterparts.
ConclusionsTaking into account that neonatal sepsis represents a significant problem on a global scale, epidemiological surveillance, antimicrobial resistance and GBS virulence at the local level could provide important knowledge about these microorganisms as well as help to improve treatment and prevent invasive infection caused by this microorganism.
Streptococcus agalactiae o estreptococos del grupo B (SGB) es el principal agente etiológico de la sepsis neonatal temprana en los países desarrollados. Este microorganismo pertenece a la microbiota del tracto gastrointestinal desde donde puede colonizar la vagina y ser transmitido verticalmente al niño antes o al nacer y posteriormente causar infección en el recién nacido. Aproximadamente el 50% de los recién nacidos de mujeres embarazadas que albergan SGB se colonizan, con 1-2% desarrollando infección neonatal temprana si no se realiza intervención preventiva. El objetivo de este estudio fue caracterizar y comparar serotipos, factores de virulencia y la resistencia a los antimicrobianos de aislamientos de SGB de mujeres embarazadas y neonatos procedentes de varios hospitales de Cataluña.
MétodosSe analizaron 242 cepas de SGB incluyendo 95 colonizadoras y 68 cepas patógenas aisladas de mujeres embarazadas y 79 cepas aisladas de neonatos con sepsis para determinar serotipo, virulencia y resistencia antimicrobiana.
ResultadosLa distribución de los serotipos fue diferente entre los 3 grupos, siendo los serotipos Ia y II significativamente más frecuentes entre las cepas colonizadoras (p=0,001 y 0,012, respectivamente). Los factores de virulencia bca y scpB fueron significativamente más frecuentes entre las cepas neonatales que entre las patógenas o colonizadoras (p=0,0001 y 0,002, respectivamente). Las cepas patógenas fueron significativamente más resistentes a eritromicina, clindamicina y azitromicina que las no patógenas.
ConclusionesTeniendo en cuenta que la sepsis neonatal es un problema importante a nivel mundial, la vigilancia de la epidemiología, la resistencia a los antimicrobianos y la virulencia del SGB a nivel local podría proporcionar un gran conocimiento de estos microorganismos y ayudar a mejorar el tratamiento y la prevención de la infección invasiva causada por este microorganismo.
Streptococcus agalactiae (group B streptococcus [GBS]) is an opportunistic pathogen that colonizes the gastrointestinal and genitourinary tract of healthy people and is responsible for severe diseases in susceptible hosts.1 Moreover, this specie is a leading cause of invasive infection in newborns, causing early neonatal sepsis in developed countries. In Spain, it has been reported that 10–18.5% of pregnant women are colonized by GBS.2
Colonization of the vagina by GBS in pregnant women is of great importance because it increases the risk of infection, due to it can be vertically transmitted to the child before or at birth, causing infection in the newborn. Infection by GBS may arise from a prematurely ruptured amniotic membrane which becomes infected or from infected amniotic fluid (chorioamnionitis) or preterm delivery in a mother colonized by these bacteria, presenting a much higher risk of infecting the offspring due to immaturity of the immune system.3
Approximately, 50% of infants from GBS-colonized women become colonized, with 1–2% of these newborns developing early or late neonatal infection if no preventive intervention is performed.4 To avoid this enhanced risk of vertical transmission, the Center of Disease Control (CDC-1996) recommended the screening of all pregnant women at 35–37 weeks of gestation to determine possible GBS colonization.5 Penicillin and ampicillin are the antibiotics of choice for treatment or intrapartum prophylaxis of GBS, followed by first-generation cephalosporins. In the case of patients allergic to β-lactams, clindamycin, erythromycin and vancomycin are the antimicrobials of choice.6,7
When implemented, the use of these prophylactic measures results in a decrease in the incidence of infection by GBS.8 In Spain, an example of the success of these prophylactic measures was demonstrated in a study carried out in 10 hospitals in Barcelona (Spain). It was found that the incidence of GBS as a cause of neonatal sepsis was reduced from 1.92/1000 newborns in 1994 to 0.26/1000 newborns in 2001 (p<0.001).2
Ten GBS serotypes have been classified (Ia, Ib and II–IX), according to recognized capsular polysaccharides that are considered to be a major virulence factor in invasive disease caused by this microorganism.10 Serotypes Ia, Ib, III and V are the most frequently involved in invasive infection.11 Additionally, GBS can develop different pathogenic mechanisms that allow colonization and invasion in different niches within the host being able to avoid and suppress the host immune response.
The aim of this study was to characterize and compare the serotype, virulence factors and antimicrobial resistance of GBS isolates from pregnant women and newborns from several hospitals in Catalonia.
Materials and methodsClinical sample and bacterial isolatesTwo hundred forty-two S. agalactiae isolates were collected from 242 patients from five Catalan hospitals (Hospital Clínic, Vall d’Hebron, San Joan de Déu, Parc Taulí and CatLab) from 2010 to 2016. Among these isolates, 95 were obtained from vaginal, rectal–vaginal and endocervical swabs from asymptomatic pregnant women at 35–37 weeks of gestation and categorized as colonizing; 68 were obtained from urine, blood, amniotic fluid, and endometrial and placental samples from symptomatic women and categorized as pathogenic isolates; and 79 isolates were isolated from blood, cerebrospinal and amniotic fluid samples of newborns with neonatal sepsis. Samples arriving to the different laboratories of Microbiology were cultured in Granada agar and selective Todd-Hewwitt broth specific for GBS isolation. Suspected colonies were confirmed by MALDI-TOF.
SerotypingDetermination of the capsular type or serotype was carried out by a multiplex-PCR using a set of primers described previously.4
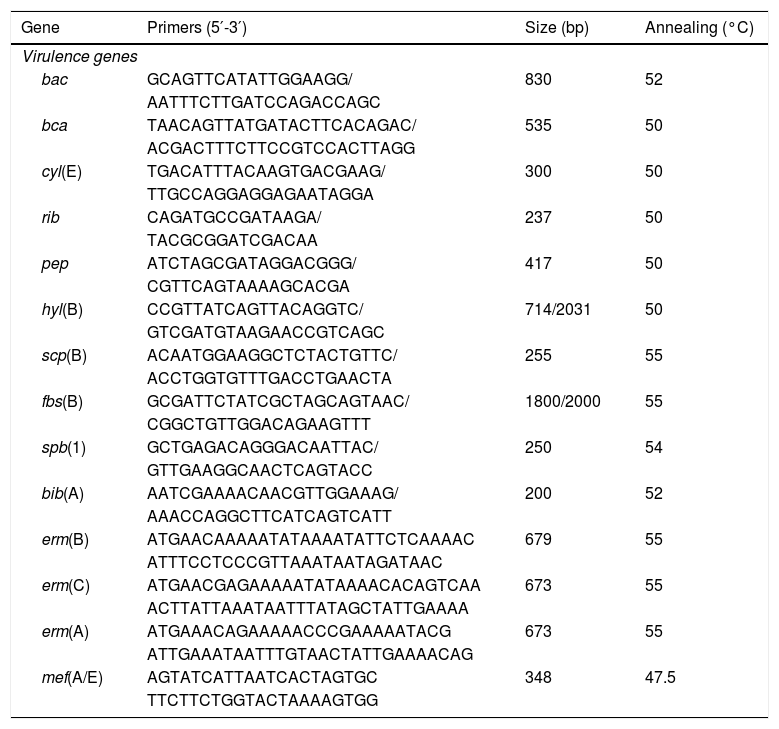
Virulence profileDifferent virulence determinants, including bac (encoding alpha subunit of the C protein), bca (encoding beta subunit of the C protein), cyl(E) (encoding cytolysin–hemolysin), rib (Alp family protein Rib), hyl(B) (encoding hyaluronidase protein), pep(B) (encoding oligopeptidase protein), scp(B) (encoding an invasin), fbs(B) (encoding fibrinogen-binding protein mediating invasivity), spb(1) (encoding a protein involved in invasivity and adherence), and bib(A) (encoding an adhesin) were investigated by PCR using specific primers (Table 1).
Primers used in the study.
| Gene | Primers (5′-3′) | Size (bp) | Annealing (°C) |
|---|---|---|---|
| Virulence genes | |||
| bac | GCAGTTCATATTGGAAGG/ | 830 | 52 |
| AATTTCTTGATCCAGACCAGC | |||
| bca | TAACAGTTATGATACTTCACAGAC/ | 535 | 50 |
| ACGACTTTCTTCCGTCCACTTAGG | |||
| cyl(E) | TGACATTTACAAGTGACGAAG/ | 300 | 50 |
| TTGCCAGGAGGAGAATAGGA | |||
| rib | CAGATGCCGATAAGA/ | 237 | 50 |
| TACGCGGATCGACAA | |||
| pep | ATCTAGCGATAGGACGGG/ | 417 | 50 |
| CGTTCAGTAAAAGCACGA | |||
| hyl(B) | CCGTTATCAGTTACAGGTC/ | 714/2031 | 50 |
| GTCGATGTAAGAACCGTCAGC | |||
| scp(B) | ACAATGGAAGGCTCTACTGTTC/ | 255 | 55 |
| ACCTGGTGTTTGACCTGAACTA | |||
| fbs(B) | GCGATTCTATCGCTAGCAGTAAC/ | 1800/2000 | 55 |
| CGGCTGTTGGACAGAAGTTT | |||
| spb(1) | GCTGAGACAGGGACAATTAC/ | 250 | 54 |
| GTTGAAGGCAACTCAGTACC | |||
| bib(A) | AATCGAAAACAACGTTGGAAAG/ | 200 | 52 |
| AAACCAGGCTTCATCAGTCATT | |||
| erm(B) | ATGAACAAAAATATAAAATATTCTCAAAAC | 679 | 55 |
| ATTTCCTCCCGTTAAATAATAGATAAC | |||
| erm(C) | ATGAACGAGAAAAATATAAAACACAGTCAA | 673 | 55 |
| ACTTATTAAATAATTTATAGCTATTGAAAA | |||
| erm(A) | ATGAAACAGAAAAACCCGAAAAATACG | 673 | 55 |
| ATTGAAATAATTTGTAACTATTGAAAACAG | |||
| mef(A/E) | AGTATCATTAATCACTAGTGC | 348 | 47.5 |
| TTCTTCTGGTACTAAAAGTGG | |||
The minimal inhibitory concentrations (MICs) were determined using the E-test method according to the protocols established by CLSI.12 The antibiotics tested were: erythromycin, clindamycin, azythromycin, ampicillin, penicillin G and vancomycin. The MLSB resistance phenotype (macrolide–lincosamide–streptogramin B) of the isolates was determined by the double disc diffusion test as described previously.12 The strain Streptococcus pneumoniae ATCC 49619 was used as the quality control.
PCR determination of the macrolide resistance phenotypeThe presence of the erm(B), erm(C), erm(A) (Subclass erm(TR)) and mef(A/E) genes was determined by PCR amplification using the primers described in Table 1. The PCR products were visualized by electrophoresis on 1% agarose gels and sent for sequencing to Beckman Coulter Genomics (United Kingdom).
Statistical analysisData related to serotypes, virulence and antimicrobial susceptibility of the three study groups were analyzed using contingency tables and Chi-square (χ2) tests. A p value <0.05 was considered statistically significant. The analyses were carried out with the StataCorp. 2015. Stata Statistical Software: Release 14. College Station, TX: StataCorp LP. Correlations of all variables have been determining by the Pearson's correlation coefficient.
ResultsTwo hundred forty-two GBS isolates were included in the study. The isolates were divided into three groups: colonizers 95/242 (39.3%) (collected from asymptomatic pregnant women), pathogenic 68/242 (28.1%) (collected from symptomatic pregnant women), and neonatal 79/242 (32.6%) (collected from infected neonates).
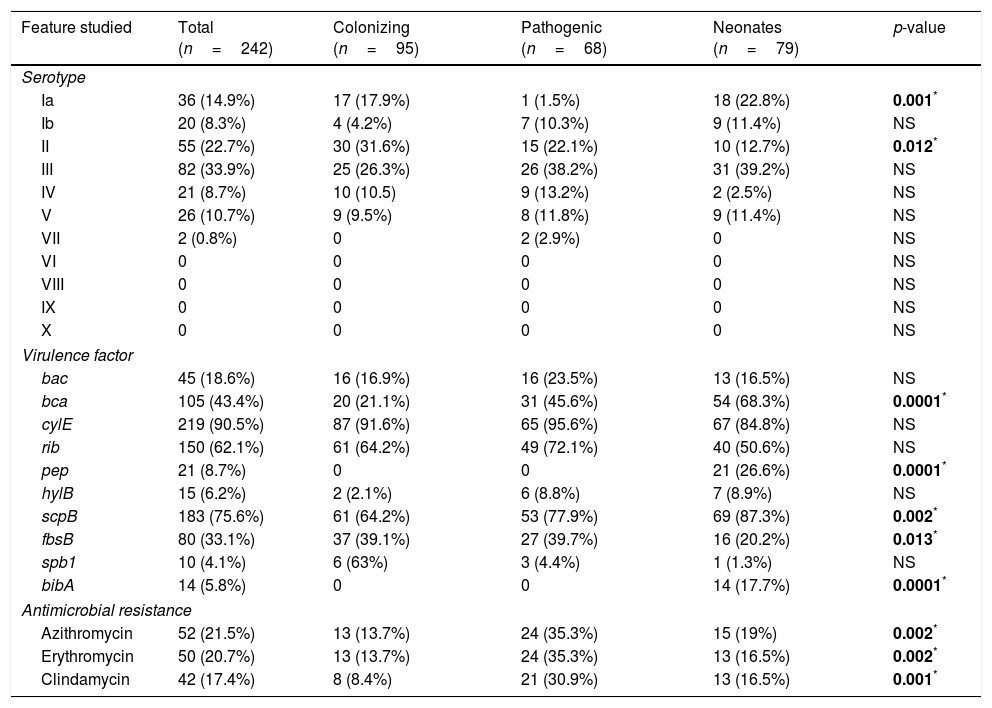
Serotype III was the most prevalent (33.9%) followed by serotypes II, Ia, V, IV, Ib, and VII (22.7%, 14.9%, 10.7%, 8.7%, 8.3% and 0.8% of isolates, respectively). However, serotypes VI, VIII, IX and X were not detected (Table 2). Among the colonizing isolates, the most frequent serotypes found were serotype II (31.6%), III (26.3%) and Ia (17.9%), while serotypes III, II and IV (38.2%, 22.1% and 13.2%, respectively) were the most prevalent among the pathogenic isolates. In the case of the neonatal isolates, serotypes III, Ia and II (39.2%, 22.8% and 12.7%, respectively) were the most frequently found. Serotype Ia was significantly more frequent among colonizing and neonatal isolates than among pathogenic isolates (p=0.001). On the other hand, serotype II was significantly more frequent among colonizing isolates (p=0.012).
Distribution of serotype, virulence factor and antimicrobial resistance of 242 GBS studied.
| Feature studied | Total (n=242) | Colonizing (n=95) | Pathogenic (n=68) | Neonates (n=79) | p-value |
|---|---|---|---|---|---|
| Serotype | |||||
| Ia | 36 (14.9%) | 17 (17.9%) | 1 (1.5%) | 18 (22.8%) | 0.001* |
| Ib | 20 (8.3%) | 4 (4.2%) | 7 (10.3%) | 9 (11.4%) | NS |
| II | 55 (22.7%) | 30 (31.6%) | 15 (22.1%) | 10 (12.7%) | 0.012* |
| III | 82 (33.9%) | 25 (26.3%) | 26 (38.2%) | 31 (39.2%) | NS |
| IV | 21 (8.7%) | 10 (10.5) | 9 (13.2%) | 2 (2.5%) | NS |
| V | 26 (10.7%) | 9 (9.5%) | 8 (11.8%) | 9 (11.4%) | NS |
| VII | 2 (0.8%) | 0 | 2 (2.9%) | 0 | NS |
| VI | 0 | 0 | 0 | 0 | NS |
| VIII | 0 | 0 | 0 | 0 | NS |
| IX | 0 | 0 | 0 | 0 | NS |
| X | 0 | 0 | 0 | 0 | NS |
| Virulence factor | |||||
| bac | 45 (18.6%) | 16 (16.9%) | 16 (23.5%) | 13 (16.5%) | NS |
| bca | 105 (43.4%) | 20 (21.1%) | 31 (45.6%) | 54 (68.3%) | 0.0001* |
| cylE | 219 (90.5%) | 87 (91.6%) | 65 (95.6%) | 67 (84.8%) | NS |
| rib | 150 (62.1%) | 61 (64.2%) | 49 (72.1%) | 40 (50.6%) | NS |
| pep | 21 (8.7%) | 0 | 0 | 21 (26.6%) | 0.0001* |
| hylB | 15 (6.2%) | 2 (2.1%) | 6 (8.8%) | 7 (8.9%) | NS |
| scpB | 183 (75.6%) | 61 (64.2%) | 53 (77.9%) | 69 (87.3%) | 0.002* |
| fbsB | 80 (33.1%) | 37 (39.1%) | 27 (39.7%) | 16 (20.2%) | 0.013* |
| spb1 | 10 (4.1%) | 6 (63%) | 3 (4.4%) | 1 (1.3%) | NS |
| bibA | 14 (5.8%) | 0 | 0 | 14 (17.7%) | 0.0001* |
| Antimicrobial resistance | |||||
| Azithromycin | 52 (21.5%) | 13 (13.7%) | 24 (35.3%) | 15 (19%) | 0.002* |
| Erythromycin | 50 (20.7%) | 13 (13.7%) | 24 (35.3%) | 13 (16.5%) | 0.002* |
| Clindamycin | 42 (17.4%) | 8 (8.4%) | 21 (30.9%) | 13 (16.5%) | 0.001* |
NS, not significant.
Regarding the presence of virulence factors, cyl(E) (90.5%), followed by scp(B) (75.6%), rib (62.1%) and bca (43.4%) (Table 2) were the most frequently found among the isolates. The virulence factors pep and bib(A) were only found among the neonatal isolates (26.6% and 17.7%, respectively). On the other hand, the bca and scp(B) genes were significantly more frequent among neonatal isolates (68.3% and 87.3%, p=0.0001 and 0.002, respectively) and the fbs(B) gene was more frequently found among colonizing (39.1%) and pathogenic isolates (39.7%) than among neonatal isolates (20.2%) (p=0.01).
Significant relationships were observed between the presence of several virulence determinants and serotypes. The bac gene was significantly more frequent among serotype Ib (p=0.0002), rib gene among serotype III (p=0.0001), spb(1) gene among serotype II (p=0.0001), and the bib(A) gene was significantly more frequent among serotype V (p=0.049).
All the isolates of this study were susceptible to penicillin, ampicillin and vancomycin. However, 21.5% were resistant to azithromycin (MIC >2μg/mL), 20.7% to erythromycin (MIC >1μg/mL), and 17.6% to clindamycin (MIC >1μg/mL). The erythromycin, azithromycin and clindamycin resistant isolates were significantly more frequent among the pathogenic than among the colonizing and neonatal isolates (p=0.002, 0.002 and 0.001, respectively) (Table 2).
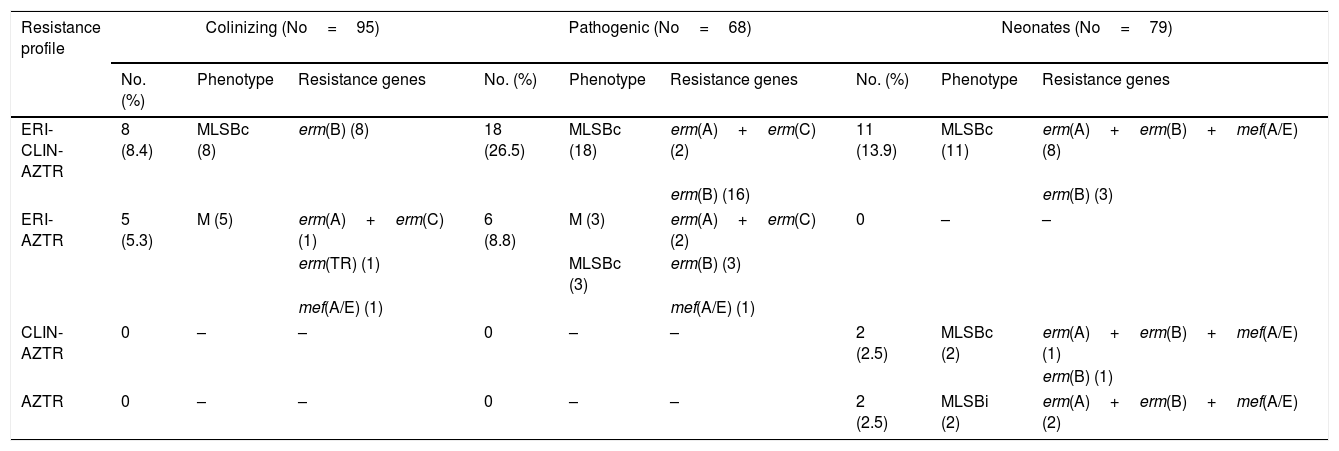
Different MLSB phenotypes were observed among the isolates; 17.8% of strains showed constitutive resistance to clindamycin (cMLSB), 3.3% showed inducible resistance (iMLSB) and 0.3% presented the M phenotype. Among the resistant isolates, the most prevalent gene found was the erm(B) gene (40/52, 77%). 12/52 (23%) unrelated isolates showed the combination of genes of erm(A)–erm(B)–mef(A/E); 5/52 (9.7%) isolates presented the erm(A)–erm(C) genes; and 2/52 (3.8%) isolates presented only the mef(A/E) gene (Table 3).
Characterization of macrolide–lincosamide resistance.
| Resistance profile | Colinizing (No=95) | Pathogenic (No=68) | Neonates (No=79) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| No. (%) | Phenotype | Resistance genes | No. (%) | Phenotype | Resistance genes | No. (%) | Phenotype | Resistance genes | |
| ERI-CLIN-AZTR | 8 (8.4) | MLSBc (8) | erm(B) (8) | 18 (26.5) | MLSBc (18) | erm(A)+erm(C) (2) | 11 (13.9) | MLSBc (11) | erm(A)+erm(B)+mef(A/E) (8) |
| erm(B) (16) | erm(B) (3) | ||||||||
| ERI-AZTR | 5 (5.3) | M (5) | erm(A)+erm(C) (1) | 6 (8.8) | M (3) | erm(A)+erm(C) (2) | 0 | – | – |
| erm(TR) (1) | MLSBc (3) | erm(B) (3) | |||||||
| mef(A/E) (1) | mef(A/E) (1) | ||||||||
| CLIN-AZTR | 0 | – | – | 0 | – | – | 2 (2.5) | MLSBc (2) | erm(A)+erm(B)+mef(A/E) (1) |
| erm(B) (1) | |||||||||
| AZTR | 0 | – | – | 0 | – | – | 2 (2.5) | MLSBi (2) | erm(A)+erm(B)+mef(A/E) (2) |
ERI, erythromycin; CLIN, clindamycin; AZTR, azithromycin.
No, number of isolates.
Correlation between serotypes, presence/absence of virulence factors and resistance to antibiotics was analyzed. No relationship was observed with antimicrobial resistance. Among colonizing isolates, the presence of bca gene was strongly associated with the presence of bac gene (Pearson's coefficient=0.80).
DiscussionS. agalactiae or group B Streptococcus (GBS) remains one of the most important etiological causes of neonatal sepsis. However, the real burden of GBS infection in newborns is likely underestimated because some cases are not adequately diagnosed, especially in developing countries.13
The present study included 242 GBS isolates collected from five hospitals in Barcelona. Among the GBS isolates studied, four serotypes (III, II, Ia and V) were the most frequent in this study, being serotype II significantly more prevalent among colonizing isolates and serotype Ia among colonizing and neonatal isolates. Our results are somewhat similar to those from other countries in which serotype III was the most frequently found among the GBS strains studied.14,15 Another recent report showed that serotype V was the most frequent in France (44.1%) followed by serotype III.16 In this sense, studies on the distribution of the GBS serotype have shown that GBS varies both geographically and over time, being serotypes Ia, III, and V more frequently involved in neonatal invasive disease, adult infections and maternal carriage.17,18
GBS presented several virulence genes that allow these microorganisms to cause infection and even to cross intact membranes and arrive to the fetus. Recent studies have been focused on analyzing the surface gene profile, but in the present work, other virulence genes such as toxins and invasins were also studied. The most frequent virulence factors found in our study were cyl(E), scp(B), rib and bca most of them related to invasion of epithelial cells.19 These results are in correlation with previous studies carried out in other geographical areas.7,20,21 In the present work, the bca (related to invasion and antophagocytosis) and scp(B) (related to invasion) genes were most prevalent among neonatal isolates, probably favoring the colonization and invasion of neonatal mucosa and blood/brain barrier.21,22 In contrast to our findings, Dutra et al.7 found the bca gene more prevalent among colonizing strains and the scp(B) gene in the 100% of the strains. The virulence factor rib related to invasiveness was more prevalent among serotype III that is associated with invasive infection and, in our study, in agreement with their higher presence in pathogenic isolates. One isolate belonging to serotype III and presenting the spb(1) and rib virulence genes formed part of the ST17 associated with neonatal invasive infections.23
In the present study, we observed a significant percentage of isolates resistant to second choice antibiotics, such as azithromycin, erythromycin and/or clindamycin. Remarkably, we found that the percentage of isolates resistant to these antibiotics was significantly higher among pathogenic than colonizing and neonatal isolates suggesting that, in contrast with other bacteria such Escherichia coli, pathogenic GBS strains are more prone to acquire antimicrobial resistance.24
Moreover, the rates of resistance to macrolides and clindamycin have also been on the rise worldwide. Domelier et al.25 and Campelo et al.9 reported 19% of resistance to erythromycin among GBS strains causing materno-fetal and neonatal disease in France and Las Palmas (Spain), respectively. On the other hand, a study carried out in Switzerland reported 30% of erythromycin resistance and 28% of clindamycin resistance,26 with a significant increase in the number of multidrug-resistant strains. However, other studies have found lower rates of isolates resistant to macrolides among pathogenic strains, being lower or equal of 4%.7,27 These differences could be due to the selective pressure caused by the antimicrobial treatments that these pregnant women could be received during gestation.
Interestingly, the resistance to macrolides and lincosamides in GBS in our study was associated with the presence of the erm(B) gene, being consistent with epidemiological studies about invasive GBS isolates collected from neonates in China and South Korea. On the other hand, the coexistence of several resistant genes such as erm(A), erm(B) and mef(A/E) (12 isolates) is remarkable in our work. These results differ from those published by other authors, who described combinations of these genes, the most common being the erm(A)/erm(B) and erm(A)/mef(A/E) genes.15,16,28
In contrast with other studies that found a significant relationship between erythromycin resistance and serotype III,29–31 this association was not present among our isolates.
In conclusion, although penicillin G and ampicillin continue to be a good treatment for GBS infections, a significant increase in the percentage of isolates resistant to macrolides and lincosamines has been observed. The fact that this increase was observed mainly among GBS causing symptomatic disease implies a serious problem for treating this type of infections. Surveillance of the epidemiology, antimicrobial resistance and virulence of GBS could provide great knowledge about these microorganisms and help to improve the treatment of infections caused by this opportunistic pathogen.
Compliance with ethical standardsThe protocols included in the manuscript are in compliance with the ethical standards.
Ethical approvalThe project related to this manuscript was approved by the Ethical Committee of the Hospital Clinic of Barcelona (CEIC). Resolution/expedient 2013/8488.
Informed consentBacterial samples were collected from the laboratories of the different hospitals. These bacterial samples were isolated from the routine samples collected for the screening under being informed by the obstetricians.
Financial supportThis material is based upon work supported by Grants PI13/00127 integrated in the Plan Nacional de I+D+I and cofunding by the “ISCIII Subdirección General de Evaluación” and the “Fondo Europeo de Desarrollo Regional (FEDER)”.
Conflict of interestThe authors declare that no conflict of interest exist.
The authors sincerely thank all the lab members who took part in this study. We are grateful to the staff of all the participating hospitals of Catalonia (Spain) for their assistance and collaboration with the isolate collections. ISGlobal is a member of the CERCA Programme, Generalitat de Catalunya.