There is an obvious need to diagnose hepatocellular carcinoma using novel non-invasive and sensitive biomarkers. Circular RNAs have recently attracted great interest as promising biomarkers and treatment targets. However, their function in hepatocellular carcinoma whose etiology related to hepatitis C has been rarely studied.
Aim of workThe current study was conducted to analyze differential expression of circ-ITCH in plasma of Egyptian HCC patients with concomitant HCV infection, compared to normal control subjects, to investigate its correlation with liver function parameters, and to determine the possible diagnostic ability of circ-ITCH in plasma as a non-invasive marker, compared to its linear counterpart.
ResultsThe results showed that the relative expression of circ-ITCH was significantly higher in the plasma of HCC patients (P<0.05). Moreover, when comparing its expression in the metastatic and non-metastatic subgroups, it was significantly higher in the non-metastatic HCC group compared to control group (P<0.05). Circ-ITCH was positively correlated with liver enzymes AST, ALT (P<0.001), also was significantly higher in HCC child C patients. To evaluate the potential diagnostic value of circ-ITCH in plasma, a ROC curve was generated, the AUC was 0.661, (95% CI: 0.5433–0.778) with a sensitivity and specificity 65% and 70% respectively.
ConclusionThe results revealed that circ-ITCH is-with no doubt-involved in the pathogenesis of HCC and its high level may be related to HCV infection, further researches in this area will certainly make great contributions in understanding. In conclusion our results suggested that circ-ITCH may be used as a noninvasive diagnostic marker and a promising therapeutic target for HCC.
Existe una necesidad obvia de diagnosticar el carcinoma hepatocelular (CHC) utilizando nuevos biomarcadores no invasivos y sensibles. Los ARN circulares han atraído recientemente un gran interés como biomarcadores prometedores y dianas de tratamiento. Sin embargo, su función en el carcinoma hepatocelular, cuya etiología está relacionada con la hepatitis C, apenas ha sido estudiada.
ObjetivoEste estudio se realizó para analizar la expresión diferencial de circ-ITCH en el plasma de pacientes egipcios con CHC con infección concomitante por VHC, en comparación con sujetos de control normales, para investigar su correlación con los parámetros de la función hepática y para determinar la posible capacidad diagnóstica de circ-ITCH en plasma como marcador no invasivo, en comparación con su contraparte lineal.
ResultadosLos resultados mostraron que la expresión relativa de circ-ITCH fue significativamente mayor en el plasma de pacientes con CHC (p<0,05). Además, al comparar su expresión en los subgrupos metastásico y no metastásico, fue significativamente mayor en el grupo de CHC no metastásico en comparación con el grupo control (p<0,05). Circ-ITCH se correlacionó positivamente con las enzimas hepáticas AST y ALT (p<0,001), y también fue significativamente mayor en pacientes con CHC infantil con VHC. Para evaluar el valor diagnóstico potencial de circ-ITCH en plasma se generó una curva ROC, el AUC fue de 0,661 (IC95%: 0,5433-0,778), con una sensibilidad y una especificidad del 65% y del 70%, respectivamente.
ConclusiónLos resultados revelaron que circ-ITCH está, sin duda, involucrado en la patogénesis del CHC, y su alto nivel puede estar relacionado con la infección por VHC, por lo que investigaciones adicionales en esta área ciertamente harán grandes contribuciones para su comprensión. En conclusión, nuestros resultados sugirieron que circ-ITCH puede usarse como un marcador de diagnóstico no invasivo y una diana terapéutica prometedora para el CHC.
Hepatocellular carcinoma (HCC) is considered the 7th most common cancer, as well as being the 2nd leading cause of cancer-related death worldwide.1,2 HCC is highly prevalent in Eastern Asia and sub-Saharan Africa regions with about 25–35 cases per 100,000 population per year, where it is associated with high hepatitis B, C viruses (HBV, HCV) prevalence.3 In Egypt HCC is a major health issue; were it is considered the fourth most common cancer, accounting for about 70.48% of all liver tumors among Egyptians,4,5 in addition to being the 2nd cause of cancer mortality in both sexes.6,7
Studies in Egypt have shown the importance of HCV infection in the etiology of liver cancer, estimated to account for 40–50% of cases of HCC.8
There are a number of factors that can predict the outcomes and prognosis of HCC. Four of the most important factors in determining survival are the severity of the accompanying liver disease, tumor size and number, extension of the tumor into other structures, and presence of metastases.9 The prognosis of HCC is thought to be poor; its 5-year survival rate is about 6.9% and this is due to its low resect-ability rate, high recurrence rate after resection and poor response to conservative treatment.6 Recent targeted therapy options provided a 5-year survival exceeding 70% for early-stage HCC and median survival of ∼1–1.5 years for symptomatic advanced-stage cases treated with systemic therapies.10 Alpha-fetoprotein (AFP), lens culinaris agglutinin A-reactive fraction of alpha-fetoprotein (AFP-L3), and des-gamma-carboxy prothrombin (DCP) have been widely used as HCC tumor markers, although they are not included in the diagnostic criteria for HCC in the guidelines of the American Association for the Study of Liver Diseases or the European Association for the Study of the Liver owing to their low sensitivity and specificity. Nevertheless, this highlights the need for minimally invasive, simple, and reliable methods for diagnosis and prognosis for HCC.11,12
Researches have shown the relations between various oncogenes or anti-oncogenes and HCC metastasis, which further led to continuous improvement in therapeutic approaches, and in understanding the exact mechanisms of HCC.
Numerous noncoding RNAs, including miRNAs and long noncoding RNAs, have been found to be deregulated in HCC patients, which were proved that they play vital regulatory roles in the cellular biological and physiological process.13,14 However, the expression profile, function, and deregulation of circRNAs in HCC remain to be identified. Interestingly, circRNAs (a class of non-coding RNAs) are recognized as a novel profile of endogenous noncoding RNAs.
Circular RNAs are a class of endogenous noncoding RNAs that form a covalently closed continuous loop without 3′ and 5′ends due to back splicing events in pre-mRNAs, and exist extensively in mammalian cells (Starke et al., 2015). They were first discovered in 1970s.15 They were considered as ‘transcriptional noise’ with little or no regulatory potential,16 produced due to splicing errors with no functions.17
Rapid development of deep sequencing and bioinformatics approaches, led to the discovery of a large number of circRNAs which were characterized with stability, prevalence, specificity and conservation.18,19 Moreover, studies revealed their abilities to sponge to miRNAs, cis-regulate parent genes, bind to proteins and encode proteins. These properties attracted great interest for circRNAs as disease biomarkers.20 CircRNAs are involved in numerous cancers such as lung cancer, gastric cancer. In addition to noncancer diseases as cardiovascular diseases, diabetes mellitus and others.20–22
Circ-ITCH is a circRNA generated from several exons of itchy E3 ubiquitin protein ligase (ITCH) gene, circbase ID (hsa_circ-0059926), it is located on chromosome 20q11.22, in a sense orientation to the protein-coding gene ITCH, and spans exons 6–13 of gene ITCH.18,20 ITCH is a member of the E3 ubiquitin ligases that regulate protein stability and immunological responses, as well as cancer progression.23 ITCH gene encodes a member of the Nedd4 family of HECT domain E3 ubiquitin ligases.24 It typically contains 4 WW domains known to associate with PPxY-containing targets, including p63, p73, and Notch1, which are usually associated with tumor formation and chemosensitivity.25 Moreover, it harbors many miRNA binding sites including those for miR-7, miR-17, miR-214, miR-128 and miR-216b, that can bind also to the 3’-UTR of ITCH, suggesting that it can function as a miRNA sponge.18,20,26
PPxY-containing targets including p63, p73, and Notch1, are usually associated with tumor formation and chemosensitivity, demonstrating the connection of ITCH to cancer biology.25 Later, some studies addressed circ-ITCH in different types of cancers as esophageal squamous cell carcinoma,26 lung cancer27 colorectal cancer,28 and epithelial ovarian cancers.29 All these studies found the same mechanism suppressing the Wnt/β-catenin pathway but by regulating different miRNAs. Moreover, Guo et al. (2017) studied circ-ITCH and its gene polymorphisms and their association with the carcinogenesis of HCC.30 It is worthy to mention that all these studies targeted tissue biopsy or cultured cell lines as their samples to detect circ-ITCH expression levels.
On the other hand, Li et al. (2020) have recently examined the expression of cir-ITCH in osteosarcoma cancer cell lines, the results were surprising, where it was significantly upregulated in osteosarcoma cancer cell lines compared to that in the human osteoblast cell line. In addition, they found that cir-ITCH could promote the migration, invasion, and growth of osteosarcoma cells, via enhancing epidermal growth factor receptor (EGFR) expression by reducing the level of miR-7.31
This had led the authors of the current study to focus on the effect of circ-ITCH in Egyptian hepatocellular carcinoma patients. Owing to our postulation that circ-ITCH may have different mechanisms in HCC which developed due to HCV infection, as studies are still scanty in this issue. The aim of this study was to analyze differential expression of circ-ITCH and its linear counterpart in the plasma of Egyptian HCC patients (HCV positive) comparing to healthy subjects, and to investigate their correlations with liver function parameters. Also, to determine the possible diagnostic ability of plasma circ-ITCH as a stable non-invasive marker compared to its linear counterpart.
Subjects and methodsSubjects81 subjects (66 males and 15females) were enrolled in this study. 51 diseased and 30 healthy controls matched in age and sex. Patients were recruited from certain outpatient's private clinic of oncology under the supervision of an oncologist from our team. All patients were previously diagnosed as HCC with concomitant HCV infection. Patients were diagnosed through CT findings, radiological assessments and clinical investigations. Provided written informed consent was obtained from patients before sample collection.
Liver function parameters, serum α-fetoprotein (AFP) levels, clinicopathological data were recorded for all patients. The Child–Pugh scoring system was performed for all patients, using grades for the following parameters of bilirubin, albumin, prothrombin time, hepatic encephalopathy, and ascites from one to three to a maximum score of 15.32 None of the patients was treated with radiotherapy or chemotherapy before plasma collection or had a previous medical history of other kind of cancers or metastatic cancer from other sites. Patients with severe disorders of major organs or a history of any other tumors were excluded.
The study was approved by the Institutional ethical board of faculty of pharmacy-Helwan university (Approval No. 1H2019, Date 17/2/2019). The study was carried out in accordance with the regulations and recommendations of the Declaration of Helsinki.
Study design and sample collection:Patients were classified into 2 subgroups: Non metastatic HCC patients (n=36), and Metastatic HCC patients (n=15).
Blood samples were withdrawn from the antecubital vein, collected into 0.5M EDTA-containing tubes for separation of plasma. Each sample was divided into several aliquots and kept at −80°C until RNA extraction.
RNA extraction and reverse transcriptionTotal RNA was extracted from 400μl plasma with miRNeasy serum/plasma total RNA purification kit (Qiagen, USA) according to the manufacturer's protocol. The extracted RNA was dissolved in RNase-free water and measured using a Nanodrop 2000 (Thermo Scientific, USA). The reverse transcription of quantified RNA was performed using Revertaid c.DNA synthesis kit (Thermofisher Scientific, USA) using random primers and according to manufacturer's instructions. C.DNA was stored in −80°C until further use in PCR.
Expression levels of circ-ITCH and ITCH m.RNA using quantitative real-time PCRA qRT-PCR assay was done using divergent primer set to determine the levels of circ-ITCH in plasma samples of patients and controls, as well as a set of convergent primers that amplifies linear RNA. QPCR assay was performed using sensiFast SYBER mix (Bioline, USA) and was monitored using Rotor-gene Q6000 detection system (Corbette Research, Australia) adjusted at 95°C for 2min first for enzyme activation, then cycling for 40 cycles at 95°C for 10s and 58°C for 45s. GAPDH was used as internal standard control. Primers for CircRNAs and the linear counterpart were synthesized by Realtime primers© (USA). The relative expression levels of circ-ITCH and ITCH m.RNA were determined using the ΔΔCt (2−ΔCt) method. The primers used for qPCR are listed in Table 1. The primer sequences have been published previously (Guo et al., 2017).30
Statistical analysisData management and statistical analysis were performed using the Statistical Package for Social Sciences (SPSS) version 24. Data were explored for normality using Shapiro–Wilk test. Categorical data were summarized as percentages.
Numerical data with normal distribution were presented as means with standard errors, comparisons between groups with respect to normally distributed numeric variables were done using the independent t-tests, ANOVA tests, while non-normal variables were described as median with interquartile range [IQR], variables were compared by non-parametric Mann–Whitney test and Kruskal–Wallis tests. For categorical variables, differences were analyzed with χ2 (chi square) test and Fisher's exact test when appropriate. P-values≤0.05 were considered significant. Correlations were done between variables by regression analysis. Receiver operating characteristic curve (ROC) was performed using Graphpad Prism version 5.0 to determine diagnostic ability of studied circ RNA.
ResultsThe clinical and demographic characteristics of all subjects included in the study were shown in Tables 2 and 3. HCC patients in this study had a significant high proportion of males (88.2%), with average age of 60 years, compared to control group (p=0.001). All patient groups were HCV positive, had significantly high liver function parameters (AST, ALT, Bilirubin and Albumin) compared to control (p=0.001). Also, AFP and INR values for blood clotting were recorded for all included groups.
Demographic characteristics of subjects included in the study.
| Parameters | HCC | Controls | P value |
|---|---|---|---|
| Age | 60.1±0.84 | 42.6±1.77 | <0.001 |
| Sex | |||
| Female | 6 (11.8%) female | 9 (30%) female | <0.001 |
| Male | 45 (88.2%) male | 21 (70%) male | |
| AST (IU/L) | 65.4±7.6 | 22.4±1.7 | <0.001 |
| ALT (IU/L) | 63.6±7.1 | 12.8±1.3 | <0.001 |
| S.Albumin (g/dl) | 3.2±0.08 | 5.1±0.2 | <0.001 |
| S.Bilirubin (mg/dl) | 2±0.3 | 1.0±0.1 | <0.001 |
| HCV coreAgHBsAg | 100% positive100% negative | 100% negative100% negative | |
| AFP (ng/ml) | 1467.6±577 | 3.21±2.65 | |
| INR | 1.31±0.05 | 0.88±0.7 |
Data are expressed as mean±SE, AST: aspartate aminotranseferase, ALT: alanine aminotransferase, AFP: α-phetoprotien, INR: international normalized ratio.
Statistics were done by independent t tests. Categorical data were compared by chi square test. P values<0.05 are considered significant.
Clinical characteristics of HCC patients.
| Clinical parameter | Count | % |
|---|---|---|
| Child stage | ||
| A | 27 | 52.9 |
| B | 18 | 35.3 |
| C | 6 | 11.8 |
| No. of lesions | ||
| One | 27 | 52.9 |
| Two | 11 | 21.6 |
| Three | 2 | 3.9 |
| Multi | 11 | 21.6 |
| P.v. invasion | ||
| No | 35 | 68.6 |
| Yes | 16 | 31.4 |
| Metastasis | ||
| No | 36 | 70.6 |
| Yes | 15 | 29.4 |
| Cirrhosis (+) | ||
| 0 | 11 | 21.6 |
| 1 | 32 | 62.7 |
| 2 | 3 | 5.9 |
| 3 | 5 | 9.8 |
| Ascites | ||
| 0 | 28 | 56.0 |
| 1 | 13 | 26.0 |
| 2 | 4 | 8.0 |
| 3 | 5 | 10.0 |
P.v: portal vein.
Cirrhosis is graded into 4 groups according to ultrasonography into 4 stages (0: no fibrosis, Grade1: mild fibrosis, Grade 2: moderate fibrosis, Grade 3: severe fibrosis or cirrhosis).
Ascites is graded into 4 stages according to clinical examinations and sonography (0: no ascites, Grade 1 (mild). ascites is only detectable by ultrasound examination, Grade 2 (moderate). ascites causing moderate symmetrical distension of the abdomen. Grade 3 (large). Ascites causing marked abdominal distension.
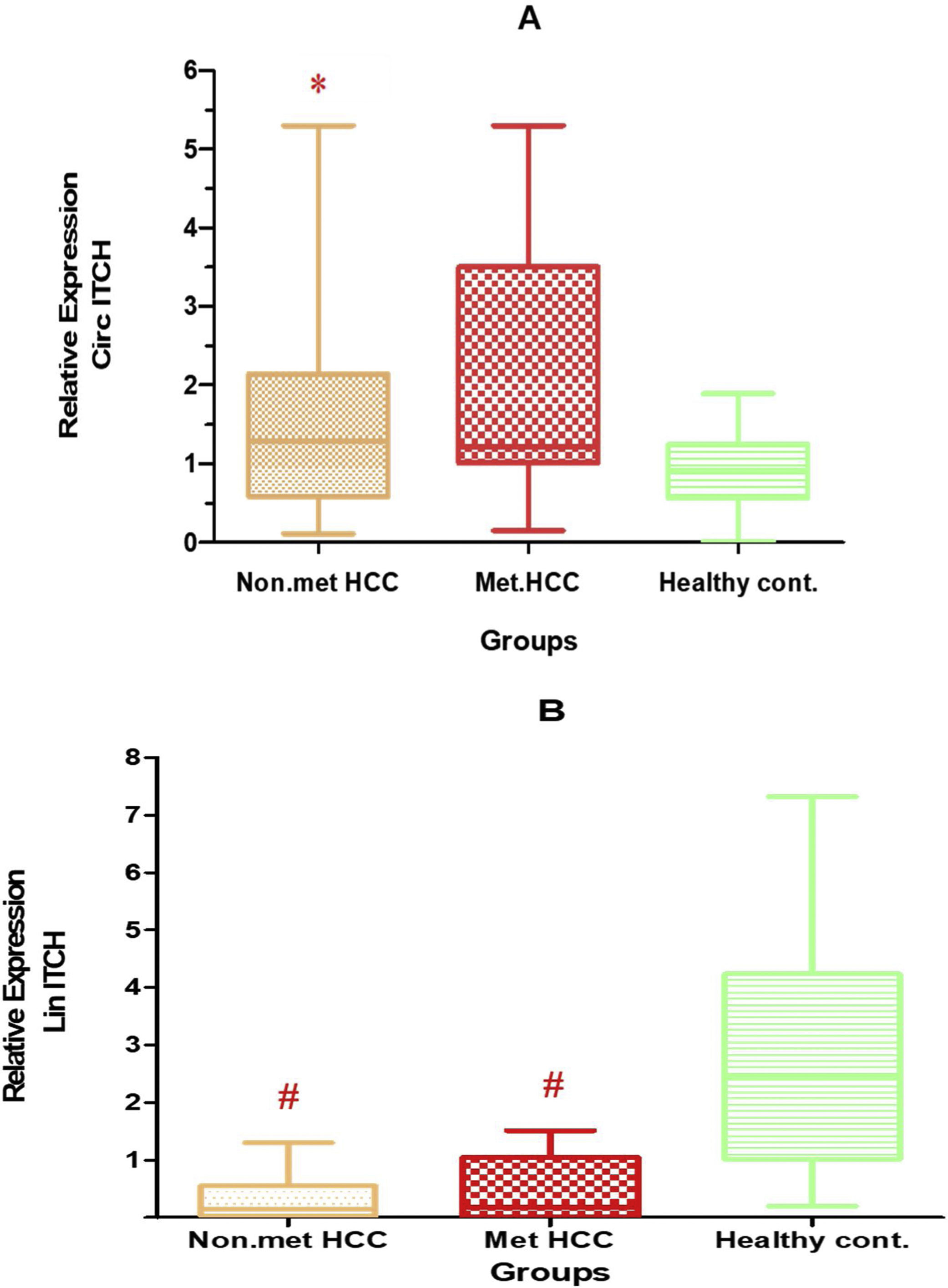
The relative expression levels of circ-ITCH in this study showed significantly higher expression levels in all HCC patients (median 1.28, range: 0.11–5.1) compared to control (median 0.9, range: 0–1.9) P<0.05, Table 4, Fig. 1A, B.
Relative expression of circ-ITCH and lin-ITCH in patients and controls.
| Total HCC | Non metastatic HCC | Metastatic HCC | Control | ||
|---|---|---|---|---|---|
| Median | Median | Median | Median | p value | |
| Circ-ITCH | 1.28a | 1.3a | 1.2 | 0.9 | 0.041 |
| lin ITCH | 0.1b | 0.1b | 0.3 | 2.2 | <0.001 |
p≤0.05 is statistically significant, analysis done by Mann–Whitney test when comparing total HCC to controls, and Kruskal–Wallis when comparing subgroups to control.
(A) Relative expression of circ-ITCH in patients and controls. (B) Relative expression of lin-ITCH in patients and controls. Statistics done by Kruskal–Wallis test. p≤0.05 is statistically significant. * p<0.05 circ-ITCH compared to control, #: p<0.01 lin-ITCH compared to control.
Stratifying HCC patients into metastatic and nonmetastatic subgroups, the relative expression in nonmetastatic group was (median 1.3, range: 0.1–5.1), while in metastatic group was (median 1.2, range: 0.1–0.3).
These values were higher than control but reach significance only in the nonmetastatic subgroup at p<0.05.
However linear-ITCH had an opposite behavior, where it showed significantly lower expression in metastatic and nonmetastatic HCC groups than control group p<0.001 (median 0.3, range: 0–1.5 and 0.1, range: 0–1.3 respectively).
Possible correlations between circular, linear-ITCH levels and clinical parameters in HCC patientsRegression analyses were done to show the correlations between the expression levels of circ-ITCH and lin ITCH with other clinicopathological features of patients with HCC such as age, sex, liver function parameters, child stage, etc. As shown in Table 5 circ-ITCH levels in the HCC patients were positively correlated with the increase in liver enzymes AST, ALT (P<0.05).
correlations between circ-ITCH, lin ITCH relative expression (RQ) and clinicopathological parameters.
| Clinicopathological parameter | Plasma circ-ITCH (RQ) | Plasma lin ITCH (RQ) | ||
|---|---|---|---|---|
| Correlation coeff. (r2) | P value | Correlation coeff. (r2) | P value | |
| Age | 0.211 | 0.06 | −0.516 | 0.001 |
| AST | 0.243 | 0.031* | −0.344 | 0.03* |
| ALT | 0.263 | 0.019* | −0.758 | <0.001** |
| s.Albumin | −0.18 | 0.343 | 0.557 | <0.001** |
| s.Bilirubin | −0.02 | 0.862 | −0.182 | 0.26 |
| AFP | 0.187 | 0.189 | 0.047 | 0.847 |
| INR | 0.103 | 0.473 | 0.047 | 0.847 |
| Plasma lin ITCH/Circ ITCH | −0.015 | 0.929 | −0.015 | 0.929 |
On the other side lin ITCH also showed a significantly negative correlation with liver enzymes AST, ALT (p=0.03, 0.001 respectively), As well as a positive correlation with serum albumin levels.
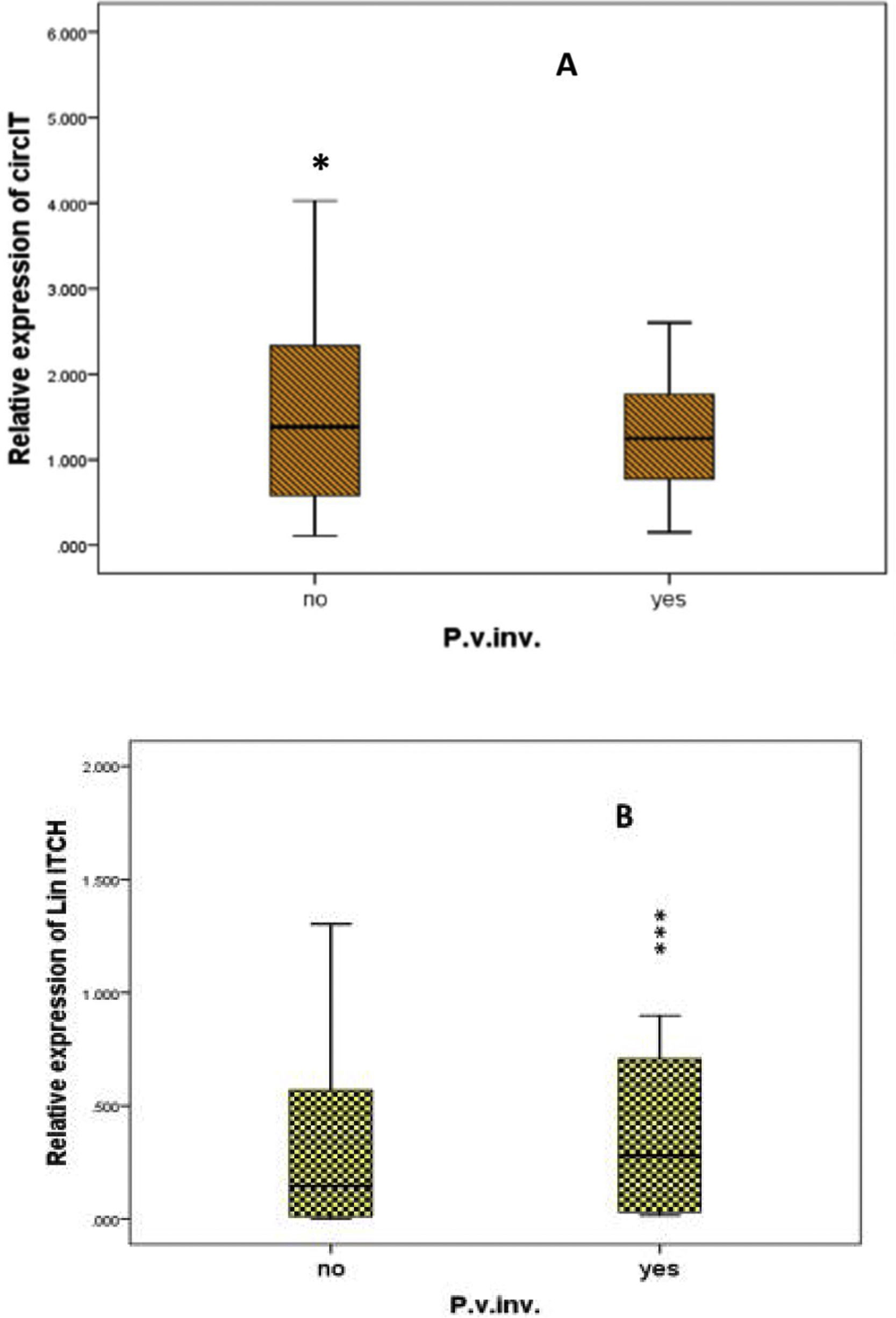
Furthermore, on comparing circ-ITCH levels in different clinical parameters as child stage, number of lesions, portal vein invasion, cirrhosis and ascites; it was found that circ-ITCH levels was significantly higher in HCC cases with no portal vein invasion (P=0.05), and was also higher in child C stage other than A and B stages, Table 6, Fig. 2A, B.
The effect of some clinicopathological parameters on the relative expression (RQ) of circ-ITCH and lin ITCH.
| Clinicopathological parameter | Plasma circ-ITCH (RQ) | Plasma lin ITCH (RQ) | ||
|---|---|---|---|---|
| Median | P value | Median | P value | |
| Sex | ||||
| Females | 0.8 | 0.132 | 0.4 | 0.679 |
| Males | 1.1 | 0.9 | ||
| No. of lesions | ||||
| One | 1.5 | 0.527 | 0.1 | 0.616 |
| Multiple | 1.1 | 0.1 | ||
| Child stage | ||||
| A | 1.5 | 0.5 | 0.105 | |
| B | 0.9 | 0.03 | ||
| C | 2.1 | 0.012* | 0.3 | |
| P.v invasion | ||||
| No | 1.4 | 0.058* | 0.1 | |
| Yes | 1.2 | 0.3 | <0.001* | |
| Cirrhosis | ||||
| No | 0.5 | 0.097 | 0.019 | 0.327 |
| Yes | 1.4 | 0.2 | ||
| Ascitis | ||||
| No | 1.4 | 0.747 | 0.4 | 0.139 |
| Yes | 1.2 | 0.1 | ||
For lin ITCH it was significantly higher in HCC cases with portal vein invasion (p<0.001).
Neither of the 2 forms of RNA had any correlation with liver ascites nor liver cirrhosis, Table 6, Fig. 2A, B.
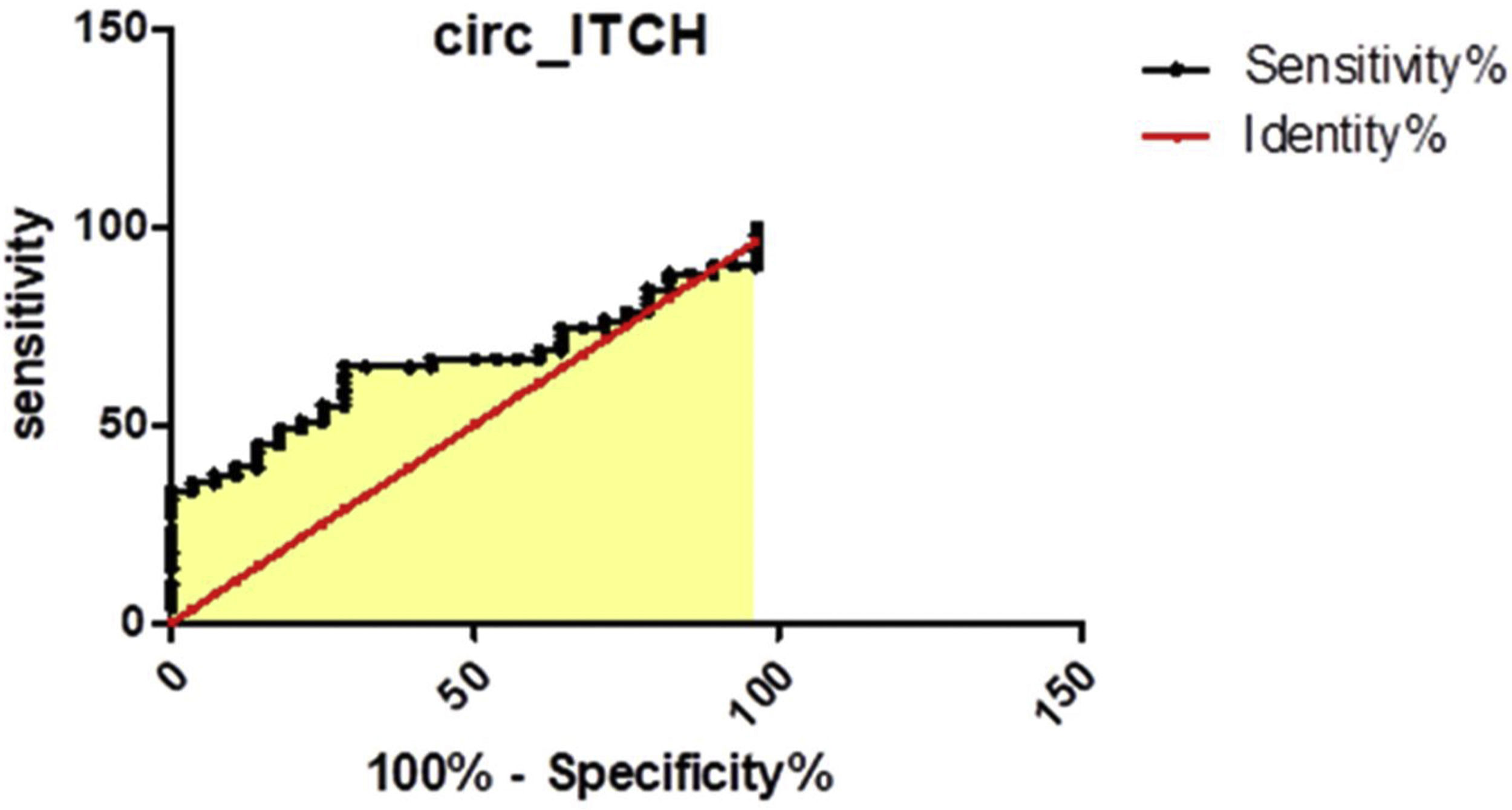
Evaluation of the potential diagnostic values of circ-ITCH and lin-ICTH:To evaluate the potential diagnostic value of the studied Circ RNA, a Receiver operator curve (ROC) curve was generated for circ-ITCH in plasma. We found that the area under the ROC curve (AUC) was 0.661, (95% CI: 0.54–0.78), Fig. 3. The sensitivity and specificity were 65% and 70% respectively at a cut off value less than or equal to 1.014.
DiscussionHCV induced HCC is one of the most dangerous cancers in Egyp.8 The shortage of appropriate biomarkers for early detection together with the lack of symptoms often results in the late diagnosis of the cancer only at higher stages, which is linked with a poorer survival.6
In this study, males constituted 88.2% of HCC patients, agreeing with the natural epidemiological classification of HCC in Egypt, where higher incidence of HCC was observed in males.8,33 Sex disparity has been also observed worldwide, where the male predominance in HCC is 2–5 times higher regardless of the etiology of HCC.34 The sex disparity in HBV/HCV-related HCC can be explained by the cross-talk between sex hormone and HBV/HCV.35
CircRNAs can be probably utilized as predictive biomarkers for cancer diagnosis. CircRNAs in HCC tumor tissues were proven to be critical factors in diagnosis, prognosis or as therapeutic agents in several reports.36–39 Despite the reported great stability of circRNAs and their availability in exosomes and some body fluids18,40; studies exploring their presence in sera or plasma of HCC patients were few. Even though some studies have found them in the peripheral blood in other metabolic diseases such as cardiovascular diseases, prediabetes and type 2 diabetes,41,42 also in cancers as breast cancer.43
Circ-ITCH had proven its association with many cancers e.g.,28,29 as well as its proven relation with carcinogenesis in HCC.30 We aimed to explore the relative expression of circ-ITCH and lin ITCH in the plasma of Egyptian HCC patients and comparing them to normal healthy controls in order to determine the possible diagnostic ability of circ-ITCH in HCC patients using the plasma as a non-invasive mean, also to compare its availability and stability in plasma compared to linear counterpart.
Itchy E3 Ubiquitin Protein Ligase protein is a protein coding gene which regulates the stability of both transmembrane receptors and intracellular substrates through canonical ubiquitylation, driving them to lysosomal and proteasomal degradation.44 According to previous research, ITCH protein can promote the ubiquitination and degradation of phosphorylated Dvl2 (Segment polarity protein disheveled homolog) and, therefore, inhibit canonical Wnt signaling. Where aberrant regulation of the Wnt signaling pathway has emerged as a prevalent theme in cancer biology.45 studies have suggested that miRNAs can bind to the 3′-UTR of ITCH mRNA to decrease its expression.46 Circ-ITCH acts as a miRNA sponge and increases the level of ITCH m-RNA, which is involved in the Wnt/β-catenin pathway.26
Therefore, circ-ITCH is suggested to play an anti-tumor role by controlling miRNA activity, and this explains the down regulation of circ-ITCH in many cancers which were studied in several studies mentioned before.26–30 They all denoted that the inhibition effects of cir-ITCH on cell growth may be related to its cooperation with Dvl2 to suppress the Wnt/β-catenin pathway.
Surprisingly, the results of the present study were in contrast to those findings, as circ-ITCH was up regulated in the plasma of HCC patients particularly the non-metastatic patients while its linear counterpart lin ITCH had an opposite behavior, where it showed significantly lower expression levels in metastatic and non-metastatic HCC groups than control group.
To our knowledge, our study was one of the few studies investigating the presence of circ-ITCH in plasma of HCC patients developed due to HCV infection.
The conflicting results between the current study and what has been published in literature might be attributed to a number of factors that impact the expression pattern of circ-ITCH in different studies. Such factors include: Firstly; the type of specimen which is a major factor since our study was specifically in plasma while others were focused on the tumor tissue specimen, or cultured cell lines.
Simply we can assume that circ-ITCH may be decreased in liver tissues, as was stated in literature, and is pooled from liver tissues to blood due to hepatocyte injury caused by HCV infection, this may be approved with the positive correlation between circ-ITCH upregulation and high plasma levels of aminotransferases AST, ALT. Circulating RNAs may be passively released from broken cells as was explained by Turchinovich et al. (2011),47 also they can, originate from immunocytes in the tumor microenvironment,48 this can partly explain our case of increasing circ-ITCH in plasma in opposite to the linear ITCH.
Another factor is; the heterogeneity of the cancer patients, concerning the etiology of HCC, where patients selected for this study were all HCV related HCC, rather than HBV, targeted in several studies such as.49,50 In our present study, we postulated that HCV may have a different effect on circ-ITCH.
HCV core protein may be one of these substrates that can undergo ubiquitination and proteasomal degradation by ITCH protein.51 ITCH protein expression is affected by the upregulation of circ-ITCH specifically. So circ-ITCH may regulate the ubiquitination and degradation of HCV core protein by some means, and so high HCV presence may be related to the upregulation of circ-ITCH the. However, this postulation needs to be further investigated in our future studies.
On the contrary to all these previous results, a recent study done by Li et al. (2020) came in agreement with our results, where they studied the role of circ-ITCH in several osteosarcoma cell lines, they found that the expression of circ-ITCH in osteosarcoma cancer cell lines was significantly upregulated. In addition, they found that circ-ITCH could promote the migration, invasion, and growth of osteosarcoma cells. Further mechanistic studies revealed that circ-ITCH could enhance epidermal growth factor receptor (EGFR) expression by reducing the level of miR-7, proving that circ-ITCH may be a candidate oncogene in osteosarcoma and may serve as a new biomarker and therapeutic target for osteosarcoma.31
MiR-7 has been proven to play a significant role in tumorigenesis of various tumors, including HCC. It can act either as a tumor suppressor or oncogene since it can participate in distinct pathways and regulate discrete target genes in different cell types, it can also regulate drug resistance and progression in several cancers through the EGFR pathway.52–54
Fang et al. (2012) analyzed the expression of miR-7in clinical HCC tissue samples and reported a decrease in its expression in HCC cases, suggesting that it acts as a tumor suppressor, can inhibit proliferation and metastasis in HCC cells in vitro and in vivo.55 Moreover, Tarek et al. (2017) found the same results in the sera of Egyptian HCC patients.56
Low levels of mir-7 in Egyptian HCC patients56 together with the results proven by Li et al. (2020), can explain the high circ-ITCH levels found in HCC patients in our study.31 However, this postulation must be further studied through other mechanistic experiments.
The present study, also revealed unexpectedly, a negative correlation between plasma circ ITCH and its linear form, opposing to what stated in other studies who stated a positive significant correlation between circular and linear counterparts in tumor tissues of colorectal and esophageal cancers.26,28
This can be explained by the extraordinary stability of circRNAs compared to their linear counterparts especially in extracellular body fluids, owing to the lack of free ends and resistance to exonucleolytic degradation.19,20 CircRNAs exhibit longer half-lives, approximately 2.5-fold longer than those of their corresponding linear counterparts on average, making it possible for continuous inspection even after long time storage of RNA samples in freezing conditions.57 Moreover, Jeck et al. (2013) stated that circRNAs expression levels can be 10-fold or higher compared to their linear isomers,18 also Li et al. (2020) stated the same results when measuring lin ITCH in osteosarcoma cell lines, they stated that the linear ITCH messenger RNA was degraded by RNase and was not detected.40
Concerning correlations between circ-ITCH and clinicopathological parameters in the current study; a significant positive correlation was found between circ-ITCH levels and liver enzymes AST, ALT in the HCC patients, this can be partly explained by tissue injury which was caused by HCV infection, which leads to the release of such enzymes as well as circ-ITCH. The only study that evaluated circ-ITCH in HCC was Guo et al.(2017),30 but they didn’t state any possible correlations between circ-ITCH and any of the clinicopathological parameters included in their study.
In the current study the increase in the levels of Circ-ITCH in the non-metastatic patients as well as in child C stage group may indicate that circ-ITCH may be related to inflammation and fibrosis. Meanwhile these assumptions may be further investigated in future mechanistic analyses.
The sensitivity and specificity of circ-ITCH in the current study were 65% and 70% respectively which is fair good. Where studies demonstrated that the sensitivity of serum AFP is 59%, AFP-L3 52%, and DCP 74%,58 however AFP is the most specific 80–94% therefore it is the most widely used tumor biomarker currently available for HCC detection, but its low sensitivity beside its well-known drawbacks, having false-positive results limits its use.11,12
In summary, our results have shown a significant higher expression level of circ-ITCH in plasma of Egyptian HCC patients related to HCV infection, compared to healthy controls. In a manner which is opposite to linear-ITCH which was lower in both metastatic and nonmetastatic HCC groups than control group, owing to outstanding stability of circ RNAs in body fluids. The mechanism and causes of this overexpression are still unknown. Circ-ITCH levels were positively correlated with liver enzymes. Instantly there is no doubt that circ-ITCH are involved in the pathogenesis of HCC. Further studies must be done to investigate the mechanism of increased level of Circ-ITCH in HCC. Also to recognize it as candidate promising therapeutic target for HCC.
Authors’ contributionsWalaa M.N. performed the experiments, analysis and was a major contributor in writing the manuscript. Dr. Sahar A.A. also performed some experiments and participated in the data analysis. Dr. Mamdouh E.S. collected the clinical information of all patients. Dr. Fathia Z. made substantial amendments to the manuscript, made significant contributions to conception, design, and provided final approval for the study to be published. All authors read and approved the final manuscript.
Conflict of interestsThe authors declare that they have no conflict of interest.
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors. I want to describe my gratitude and thanks to all my faculty colleagues – Faculty of pharmacy, especially my head of department – department of Biochemistry and Molecular Biology, and all other members for support and help.