Genotypic distribution and epidemiology of HCV infection in Western Europe countries has changed over the last decades.
AimTo establish the local genotypic profile and characterize the associated demographic variables.
Material and methodAll the genotyping from 1988 to 2015 were considered. Associated demographic variables were included in logistic regression models. Genotyping was carried out with updated commercial kits.
ResultsGenotype 1b was the most prevalent (42.4%) followed by 1a (22.5%), 3 (18.6%), 4 (10.6%) and 2 (4.6%). The prevalence of 1a was higher in males, in patients younger than 45 and in intravenous drug users (IDU). 1b was more frequent in older than 45, with transfusion-associated and parenteral/nosocomial infections and in immigrants from Eastern Europe. Genotype 2 was highly prevalent in the postransfusional route (54.9%). Genotype 3 prevalence was high in males, in patients younger than 45, in IDU (69.3%) and in Asian and Eastern European immigrants. Genotype 4 was high in males, in patients younger than 45, and in IDU (63.5%). 1a, 3, 4 were the most prevalent genotypes in HIV-coinfected patients. There was a significant decline in genotype 1b and an increase in genotypes 3 and 4 over time.
ConclusionsThere has been a decline of genotype 1b, associated with transfusion or parenteral/nosocomial infections, and increases in the prevalence of genotypes 1a, 3 and 4 associated with male gender and IDU, now the most prevalent infection route. Immigration contributed with genotype 2 infections from Africa and genotype 1b and 3 infections from Eastern Europe and Asia.
La distribución genotípica y la epidemiología de la infección por el VHC en los países de Europa Occidental ha variado en las últimas décadas.
ObjetivoEstablecer el perfil genotípico local y distinguir las variables demográficas asociadas.
Material y métodoSe han tenido en cuenta todas las genotipificaciones desde 1988 a 2015. Las variables demográficas asociadas se incluyeron en modelos de regresión logística. La genotipificación se realizó con kits comerciales actualizados.
ResultadosEl genotipo 1b fue el más prevalente (42,4%), seguido por 1a (22,5%), 3 (18,6%), 4 (10,6%) y 2 (4,6%). La prevalencia de 1a fue mayor en varones, en pacientes menores de 45 años y en consumidores de drogas por vía intravenosa (CDVI). El genotipo 1b fue más frecuente en pacientes mayores de 45 años, con infecciones relacionadas con la transfusión y de tipo parenteral/nosocomial, y en inmigrantes de Europa Oriental. El genotipo 2 fue muy prevalente en la vía postransfusional (54,9%). La prevalencia del genotipo 3 fue elevada en varones, en pacientes menores de 45 años, en CDVI (69,3%) y en inmigrantes asiáticos y de Europa Oriental. El genotipo 4 fue elevado en varones, en pacientes menores de 45 años y en CDVI (63,5%). Los genotipos 1a, 3 y 4 fueron los más prevalentes en pacientes coinfectados con el VIH. Hubo una disminución considerable del genotipo 1b y un aumento en los genotipos 3 y 4 en el tiempo.
ConclusionesSe ha producido una disminución del genotipo 1b, relacionado con transfusiones o infecciones parenterales/nosocomiales, y un aumento en la prevalencia de los genotipos 1a, 3 y 4, relacionados con el sexo masculino y los CDVI, que actualmente son la vía de infección más prevalente. La inmigración contribuyó con infecciones del genotipo 2 de África e infecciones de los genotipos 1b y 3 de Europa Oriental y Asia.
Hepatitis C virus (HCV) infection constitutes a severe public health problem with approximately 80 (64–103) million viremic patients worldwide,1 and a natural history leading to liver cirrhosis which entails a risk of hepatocellular carcinoma between 1 and 5% per year.2,3 HCV shows a great genetic variability due to its high replication rate and the reduced capacity of the viral polymerase to correct the errors of the genome that appear during replication. This variability has resulted, in addition to quasi species in every infected subject, in seven viral genotypes (denoted 1 to 7) and more than 67 subtypes (denoted alphabetically a, b, c, etc.).4 The majority of infections are caused by genotypes 1a, 1b, 2, 3 (almost exclusively 3a) and 4, while infections with genotypes 5, 6 and 7 are less frequent or limited to specific geographic areas.1,5
The viral genotype has been determinant in the therapeutic guidelines of chronic HCV infection with pegylated-interferon plus ribavirin6 and with first-generation protease inhibitors.7,8 Although its usefulness has been greatly reduced by the introduction of new pangenotypic oral treatments,9–11 genotype determination remains crucial due to the marked geographical differences in the genotypic profiles which may be affected by migration trends and by changes in the routes of infection.1,5,12–16
At the end of World War II there was an explosive spread of genotype 1b infections associated with the mass implementation of blood transfusions and unsafe health practices in Western countries.15 Almost simultaneously there were a significant spread of genotype 1a linked to unsafe health practices and the rapid expansion of intravenous drug use (IDU).15 These changes paralleled with a marked increase in genotype 3a infection from the Asian continent associated with the heroin trade and IDU16 and migratory movements of certain populations.17 The spread of genotype 4, endemic in the Middle East, Egypt and Central Africa, to Europe has had different epidemiological settings. While in Egypt genotype 4a spread was propagated by mass parenteral treatment campaigns for the control of schistosomiasis during the twentieth century,18 the spread of other subtypes of genotype 4 in the developed world, especially in Europe, is much more recent and is linked to demographic movements and, above all, to IDU.19,20
The aim of this study was to establish the genotypic profile of HCV infection in the study area of Girona, Catalonia, north-eastern Spain, and to record the changes that have occurred over more than three decades.
MethodsStudy designDescriptive, retrospective cohort study designed to define its characteristics and to identify independent variables. The main objective was to define the genotypic profile of HCV infection in the study area and its evolution over the period 1980–2015. The secondary aim was to identify demographic variables associated with the dependent variables which were the viral genotypes.
PopulationAll the HCV genotype determinations performed at the Department of Clinical Laboratory of the Hospital de Girona Doctor Josep Trueta (HdGJT) from its introduction in June 1998 until 31 December 2015 were included. HdGJT is a second level referral hospital serving a population of 753,054 inhabitants in 2015. Genotype determinations were mainly investigated in chronic hepatitis C outpatients seen at the Departments of Digestive Diseases, Internal Medicine and Infectious Diseases, usually in the initial specialized assessment and whenever antiviral treatment was considered.
VariablesDemographic data were collected from all patients who had a genotype study and in those which had more than one determination the consistency of the results was confirmed. Individual records included age, gender and year of the patient's first specialized evaluation or, if there was no patient follow-up, when the sample had been obtained, HCV genotype, probable route of infection, polymorphism rs12979860, native vs immigrant status, geographical area of origin and HIV coinfection. Age was used as a categorical variable, with age groups (<20; 20–29; 30–39; 40–49; 50–59 and ≥60) in the descriptive analysis or as ≤45 vs >45 year-old in the analysis of demographic factors associated with genotypes. The 45-year-old cut-off value was chosen after considering the different genotype-associated median ages. The following routes of infection were considered: sporadic (when no route of infection was ascertained after questioning the patient), IDU, transfusion-associated, parenteral/nosocomial and undetermined. The occasional patients who referred more than one possible route of infection were classified as having the most hierarchically likely route (IDU > transfusional > parenteral/nosocomial > sporadic). As for the geographical origin patients were included in one of the following regions: Western Europe, Eastern Europe and the Caucasus, Africa, Latin America and Asia.
The data were collected retrospectively from June 1998 until 31 December 2015. The time periods considered for analyzing changes over time were 1980–1994, 1995–2005 and 2006–2015, giving priority to periods of similar length rather than to sample size. The study of the rs12979860 polymorphism was performed from 2012 onwards, only when treatment was considered and with the consent of patients.
During data collection, patients’ anonymity was ensured at all times. The authors who processed and analyzed the digital information (DAF and MJFI) did not have access either to the patients or to the prescription of antiviral therapies.
Laboratory techniquesThe viral genome was identified by extraction of viral RNA, amplification by RT-PCR and subsequent reverse hybridization. The reverse hybridization techniques in the 5′ non-coding region (5′NC) of the viral genome were INNO-LIPA HCV II (Innogenetics, Brussels), from June 1998 to July 2006 and LINEAR ARRAY HCV Genotyping Test, Roche) from July 2006 to October 2014. The lower sustained virological response (SVR) rates with first-generation protease inhibitors treatments in patients infected with genotype 1a,21,22 and the need to identify the subtypes of genotype 1, led to a change from LINEAR ARRAY to Versant HCV genotype 2.0 Assay (LiPA), (Siemens), reverse hybridization in the 5’NC regions and core, in October 2014. Patients with genotype 1 infection who did not achieve SVR during the LINEAR ARRAY period and were not lost to follow-up were subtyped with Versant-Siemens. The genotypes studied were 1, 1a, 1b, 2, 3, 3a, 4, 5 and 6 in the Simmonds classification.4
Statistical analysisCategorical variables were summarized as absolute values and percentages and quantitative variables as median and interquartile range (IQR). Pearson chi-square or Fisher's exact test was used to analyze categorical variables. Cochran-Mantel-Haenszel test was used to test for linear trend using row or column score. Quantitative variables were compared by non-parametric Mann–Whitney U test or Kruskall–Wallis test, as appropriate.
Unconditional binary logistic regression was performed to obtain odds ratios (ORs) and their 95% confidence intervals (95%CIs) for the relationship between each HCV genotype (1a, 1b, 2, 3 or 4) and socio-demographic characteristics. Univariate logistic regression was used to determine which variables showed individual associations and multivariate analyses were performed to evaluate the joint relationship of all variables that were significant in univariate analysis. Multivariate logistic regression was built using backward stepwise selection algorithm and all models were adjusted for age and gender. The Hosmer–Lemeshow test was used to assess goodness-of-fit in the models. All statistical tests were two-tailed and a p-value less than 0.05 was considered statistically significant.
Data were analyzed using SPSS software v23.0 (SPSS Inc, Chicago, IL, USA) and R Core Team (2016).
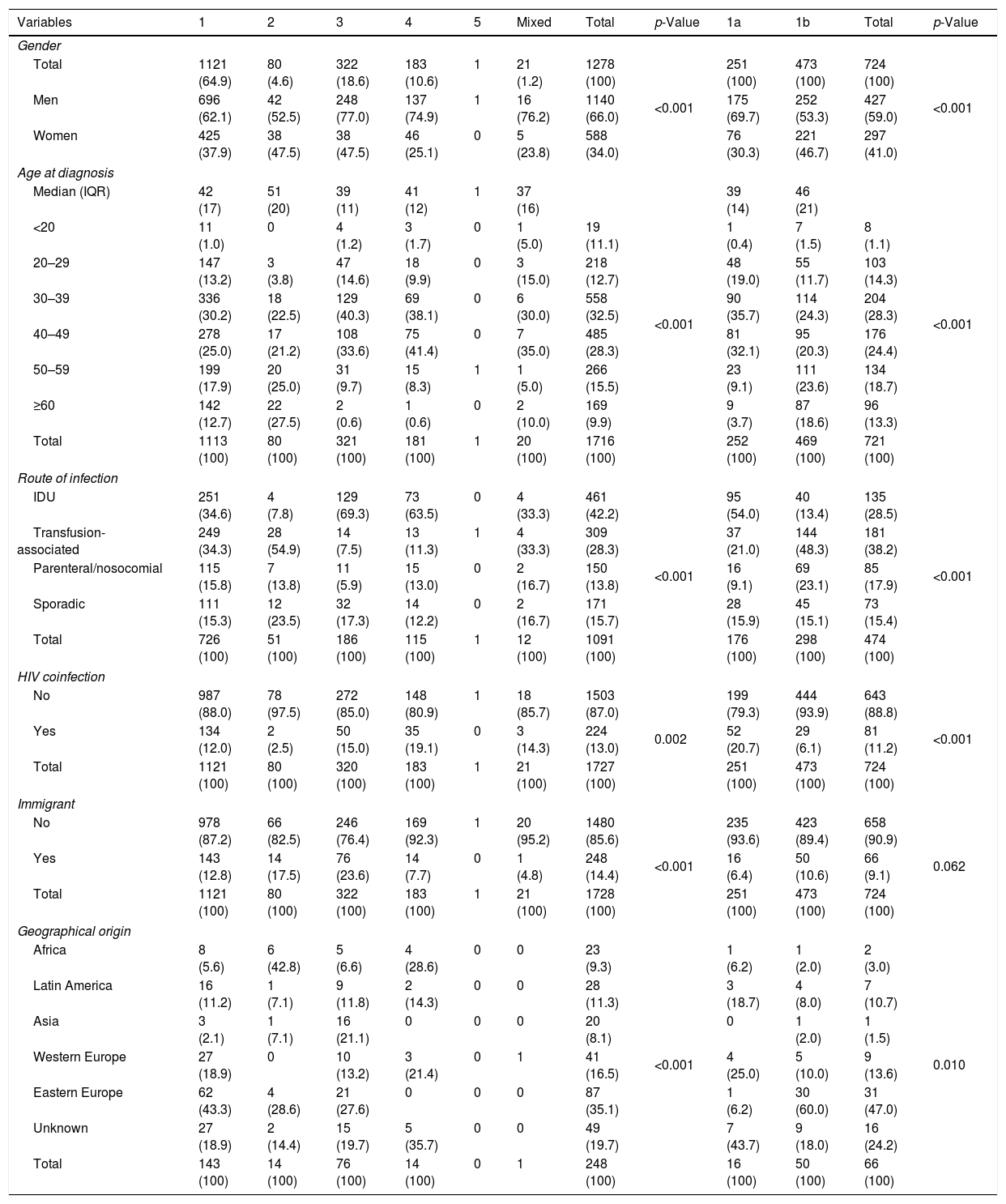
ResultsThe HCV genotype was identified in 1728 patients, 1140 men (66.0%) and 588 women (34.0%). Men were significantly younger (median age 41 years; IQR 13) than women (median 43 years, IQR 20), (p<0.001). Table 1 shows the global distribution of genotypes according to gender, age groups, route of infection, HIV coinfection and geographical origin. Genotype 1 was the most frequently found (64.9%) followed by genotype 3 (18.6%), genotype 4 (10.6%) and genotype 2 (4.6%). A mixed infection was found in 21 patients (1.2%) and genotype 5 in only one. Of the 1121 patients infected with genotype 1, the subtype was analyzed in 724. Genotype 1a infection occurred in 22.5% of patients and genotype 1b in 42.4%. All the 43 patients with genotype 3 subtypes were 3a. The study identified 21 mixed genotypes, most of them consisting of a subtype of genotype 1 plus genotypes 3 and/or 4 and rarely of genotype 2. These infections were almost exclusively present in native males with transfusion-associated or IDU and were mainly identified in the second period of the study (INNO-LIPA HCV II, Innogenetics, Brussels).
Hepatitis C virus genotype distribution according to socio-demographic characteristics.
| Variables | 1 | 2 | 3 | 4 | 5 | Mixed | Total | p-Value | 1a | 1b | Total | p-Value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gender | ||||||||||||
| Total | 1121 (64.9) | 80 (4.6) | 322 (18.6) | 183 (10.6) | 1 | 21 (1.2) | 1278 (100) | <0.001 | 251 (100) | 473 (100) | 724 (100) | <0.001 |
| Men | 696 (62.1) | 42 (52.5) | 248 (77.0) | 137 (74.9) | 1 | 16 (76.2) | 1140 (66.0) | 175 (69.7) | 252 (53.3) | 427 (59.0) | ||
| Women | 425 (37.9) | 38 (47.5) | 38 (47.5) | 46 (25.1) | 0 | 5 (23.8) | 588 (34.0) | 76 (30.3) | 221 (46.7) | 297 (41.0) | ||
| Age at diagnosis | ||||||||||||
| Median (IQR) | 42 (17) | 51 (20) | 39 (11) | 41 (12) | 1 | 37 (16) | <0.001 | 39 (14) | 46 (21) | <0.001 | ||
| <20 | 11 (1.0) | 0 | 4 (1.2) | 3 (1.7) | 0 | 1 (5.0) | 19 (11.1) | 1 (0.4) | 7 (1.5) | 8 (1.1) | ||
| 20–29 | 147 (13.2) | 3 (3.8) | 47 (14.6) | 18 (9.9) | 0 | 3 (15.0) | 218 (12.7) | 48 (19.0) | 55 (11.7) | 103 (14.3) | ||
| 30–39 | 336 (30.2) | 18 (22.5) | 129 (40.3) | 69 (38.1) | 0 | 6 (30.0) | 558 (32.5) | 90 (35.7) | 114 (24.3) | 204 (28.3) | ||
| 40–49 | 278 (25.0) | 17 (21.2) | 108 (33.6) | 75 (41.4) | 0 | 7 (35.0) | 485 (28.3) | 81 (32.1) | 95 (20.3) | 176 (24.4) | ||
| 50–59 | 199 (17.9) | 20 (25.0) | 31 (9.7) | 15 (8.3) | 1 | 1 (5.0) | 266 (15.5) | 23 (9.1) | 111 (23.6) | 134 (18.7) | ||
| ≥60 | 142 (12.7) | 22 (27.5) | 2 (0.6) | 1 (0.6) | 0 | 2 (10.0) | 169 (9.9) | 9 (3.7) | 87 (18.6) | 96 (13.3) | ||
| Total | 1113 (100) | 80 (100) | 321 (100) | 181 (100) | 1 | 20 (100) | 1716 (100) | 252 (100) | 469 (100) | 721 (100) | ||
| Route of infection | ||||||||||||
| IDU | 251 (34.6) | 4 (7.8) | 129 (69.3) | 73 (63.5) | 0 | 4 (33.3) | 461 (42.2) | <0.001 | 95 (54.0) | 40 (13.4) | 135 (28.5) | <0.001 |
| Transfusion-associated | 249 (34.3) | 28 (54.9) | 14 (7.5) | 13 (11.3) | 1 | 4 (33.3) | 309 (28.3) | 37 (21.0) | 144 (48.3) | 181 (38.2) | ||
| Parenteral/nosocomial | 115 (15.8) | 7 (13.8) | 11 (5.9) | 15 (13.0) | 0 | 2 (16.7) | 150 (13.8) | 16 (9.1) | 69 (23.1) | 85 (17.9) | ||
| Sporadic | 111 (15.3) | 12 (23.5) | 32 (17.3) | 14 (12.2) | 0 | 2 (16.7) | 171 (15.7) | 28 (15.9) | 45 (15.1) | 73 (15.4) | ||
| Total | 726 (100) | 51 (100) | 186 (100) | 115 (100) | 1 | 12 (100) | 1091 (100) | 176 (100) | 298 (100) | 474 (100) | ||
| HIV coinfection | ||||||||||||
| No | 987 (88.0) | 78 (97.5) | 272 (85.0) | 148 (80.9) | 1 | 18 (85.7) | 1503 (87.0) | 0.002 | 199 (79.3) | 444 (93.9) | 643 (88.8) | <0.001 |
| Yes | 134 (12.0) | 2 (2.5) | 50 (15.0) | 35 (19.1) | 0 | 3 (14.3) | 224 (13.0) | 52 (20.7) | 29 (6.1) | 81 (11.2) | ||
| Total | 1121 (100) | 80 (100) | 320 (100) | 183 (100) | 1 | 21 (100) | 1727 (100) | 251 (100) | 473 (100) | 724 (100) | ||
| Immigrant | ||||||||||||
| No | 978 (87.2) | 66 (82.5) | 246 (76.4) | 169 (92.3) | 1 | 20 (95.2) | 1480 (85.6) | <0.001 | 235 (93.6) | 423 (89.4) | 658 (90.9) | 0.062 |
| Yes | 143 (12.8) | 14 (17.5) | 76 (23.6) | 14 (7.7) | 0 | 1 (4.8) | 248 (14.4) | 16 (6.4) | 50 (10.6) | 66 (9.1) | ||
| Total | 1121 (100) | 80 (100) | 322 (100) | 183 (100) | 1 | 21 (100) | 1728 (100) | 251 (100) | 473 (100) | 724 (100) | ||
| Geographical origin | ||||||||||||
| Africa | 8 (5.6) | 6 (42.8) | 5 (6.6) | 4 (28.6) | 0 | 0 | 23 (9.3) | <0.001 | 1 (6.2) | 1 (2.0) | 2 (3.0) | 0.010 |
| Latin America | 16 (11.2) | 1 (7.1) | 9 (11.8) | 2 (14.3) | 0 | 0 | 28 (11.3) | 3 (18.7) | 4 (8.0) | 7 (10.7) | ||
| Asia | 3 (2.1) | 1 (7.1) | 16 (21.1) | 0 | 0 | 0 | 20 (8.1) | 0 | 1 (2.0) | 1 (1.5) | ||
| Western Europe | 27 (18.9) | 0 | 10 (13.2) | 3 (21.4) | 0 | 1 | 41 (16.5) | 4 (25.0) | 5 (10.0) | 9 (13.6) | ||
| Eastern Europe | 62 (43.3) | 4 (28.6) | 21 (27.6) | 0 | 0 | 0 | 87 (35.1) | 1 (6.2) | 30 (60.0) | 31 (47.0) | ||
| Unknown | 27 (18.9) | 2 (14.4) | 15 (19.7) | 5 (35.7) | 0 | 0 | 49 (19.7) | 7 (43.7) | 9 (18.0) | 16 (24.2) | ||
| Total | 143 (100) | 14 (100) | 76 (100) | 14 (100) | 0 | 1 | 248 (100) | 16 (100) | 50 (100) | 66 (100) | ||
[n (column %)]; IQR: interquartile range; IDU: intravenous drug use.
The genotype profile differed according to gender, with males presenting a higher prevalence of genotypes 1a (69.7%), 3 (77.0%) and 4 (74.9%) than females (p<0.005). HIV-coinfected patients had a higher prevalence of genotypes 1a, 3 and 4 and lower of genotypes 1b and 2 than non-coinfected patients (p<0.001).
The most likely route of infection was successfully established in 1091 (63.1%) out of the 1728 patients. IDU was the most frequent route of infection in this series (42.2%) followed by transfusion-associated transmission (28.3%). The IDU route was much more frequent in genotype 1a (54.0%) than in 1b (13.4%); the opposite occurred in the transfusion (21.0% vs 48.3%) and nosocomial routes (9.1% vs 23.1%), (p<0.001). There were very high rates of IDU in genotype 3 (69.3%) and genotype 4 (63.5%) infected patients and of transfusion origin in the genotype 2 (54.9%). All these differences were statistically significant (p<0.005).
This series included 248 immigrants (14.3%), significantly younger than the native patients (37; IQR 16 vs 42; IQR 16; p<0.005), of diverse geographical origin mainly from Eastern European countries (35.1%), especially from Russia, Ukraine and Romania. Other countries of Western Europe (16.5%) and Latin America (11.3%) followed suit, with a minor proportion of African (9.3%) and Asian patients (8.1%). The geographical origin could not be identified in the 19.7% of the cases. African patients had the highest rates of infection by genotype 2 (42.8%) and genotype 4 (18.6%) and Asian patients by genotype 3 (21.1%), whereas in patients from Eastern Europe and Latin American the predominant genotype was genotype 1b (60.0%) with notable rates of genotype 3 infections (27.6%). The age of the immigrant population was lower than that of the native population across all genotypes (data not shown).
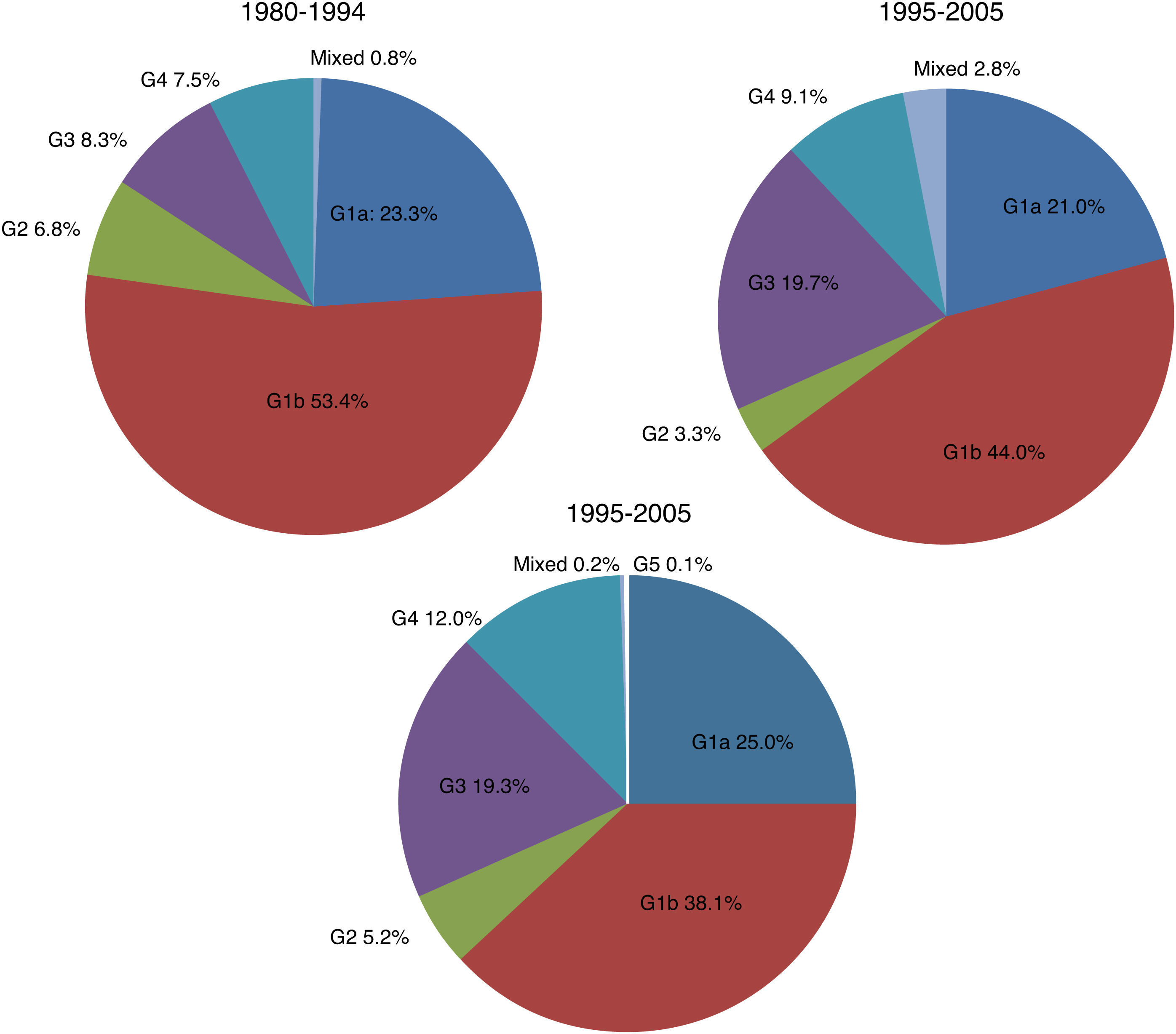
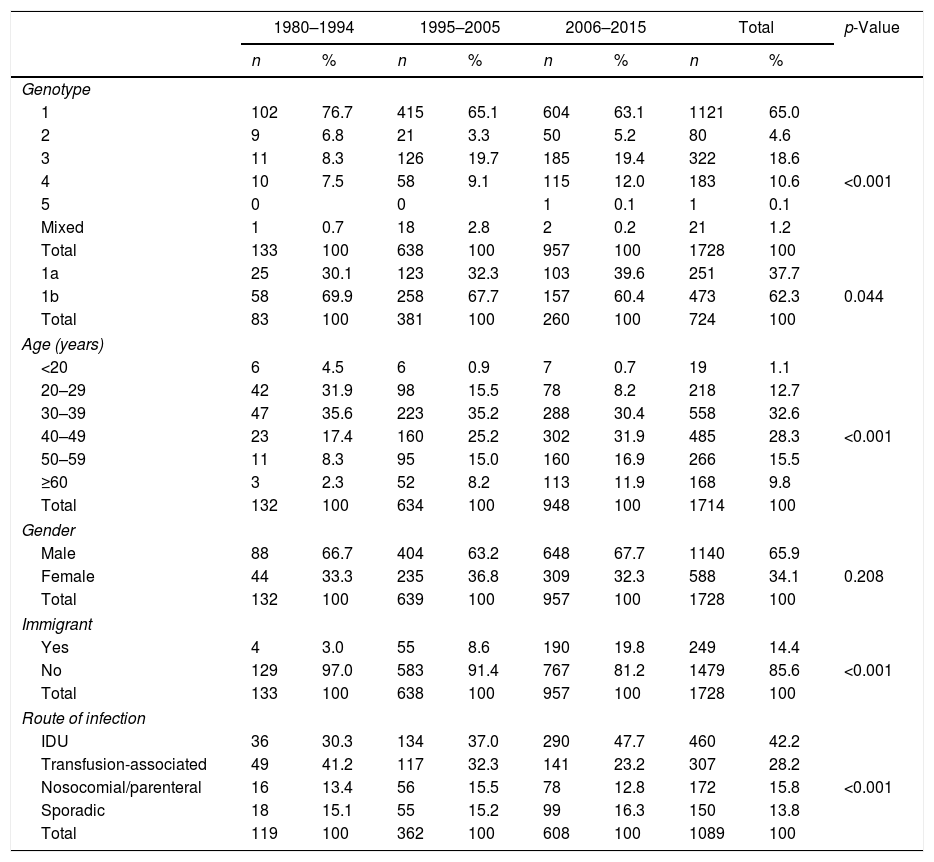
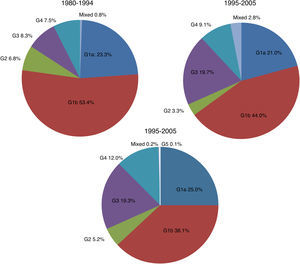
There were significant changes in the genotypic profile, age, route of infection, and geographical origin of patients throughout all the study periods (Table 2; Fig. 1). There was a steady decline in the prevalence of genotype 1 and a gradual rise in genotype 4. The prevalence of genotype 3 prevalence increased notably in the 1995–2004 period and remained stable afterward. The decrease seen in the prevalence of genotype 1 was apparently due to a fall in genotype 1b in spite of arise in genotype 1a, especially in the last period. At the same time, there was a progressive increase in the prevalence of infection acquired by IDU, a decrease in transfusion-associated infections and an increase in the percentage of immigrants. There was a significant increase in age across all genotypes throughout the three study periods. This difference did not reach statistical significance for genotype 2 (p: 0.112) probably due to the contribution of immigrants, especially from Africa, in the last period, clearly younger than their native counterparts (median age 40.5; IQR 18; p: 0.063).
Changing genotype patterns and socio-demographic variables over time [n (column (%)].
| 1980–1994 | 1995–2005 | 2006–2015 | Total | p-Value | |||||
|---|---|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | n | % | ||
| Genotype | |||||||||
| 1 | 102 | 76.7 | 415 | 65.1 | 604 | 63.1 | 1121 | 65.0 | <0.001 |
| 2 | 9 | 6.8 | 21 | 3.3 | 50 | 5.2 | 80 | 4.6 | |
| 3 | 11 | 8.3 | 126 | 19.7 | 185 | 19.4 | 322 | 18.6 | |
| 4 | 10 | 7.5 | 58 | 9.1 | 115 | 12.0 | 183 | 10.6 | |
| 5 | 0 | 0 | 1 | 0.1 | 1 | 0.1 | |||
| Mixed | 1 | 0.7 | 18 | 2.8 | 2 | 0.2 | 21 | 1.2 | |
| Total | 133 | 100 | 638 | 100 | 957 | 100 | 1728 | 100 | |
| 1a | 25 | 30.1 | 123 | 32.3 | 103 | 39.6 | 251 | 37.7 | 0.044 |
| 1b | 58 | 69.9 | 258 | 67.7 | 157 | 60.4 | 473 | 62.3 | |
| Total | 83 | 100 | 381 | 100 | 260 | 100 | 724 | 100 | |
| Age (years) | |||||||||
| <20 | 6 | 4.5 | 6 | 0.9 | 7 | 0.7 | 19 | 1.1 | <0.001 |
| 20–29 | 42 | 31.9 | 98 | 15.5 | 78 | 8.2 | 218 | 12.7 | |
| 30–39 | 47 | 35.6 | 223 | 35.2 | 288 | 30.4 | 558 | 32.6 | |
| 40–49 | 23 | 17.4 | 160 | 25.2 | 302 | 31.9 | 485 | 28.3 | |
| 50–59 | 11 | 8.3 | 95 | 15.0 | 160 | 16.9 | 266 | 15.5 | |
| ≥60 | 3 | 2.3 | 52 | 8.2 | 113 | 11.9 | 168 | 9.8 | |
| Total | 132 | 100 | 634 | 100 | 948 | 100 | 1714 | 100 | |
| Gender | |||||||||
| Male | 88 | 66.7 | 404 | 63.2 | 648 | 67.7 | 1140 | 65.9 | 0.208 |
| Female | 44 | 33.3 | 235 | 36.8 | 309 | 32.3 | 588 | 34.1 | |
| Total | 132 | 100 | 639 | 100 | 957 | 100 | 1728 | 100 | |
| Immigrant | |||||||||
| Yes | 4 | 3.0 | 55 | 8.6 | 190 | 19.8 | 249 | 14.4 | <0.001 |
| No | 129 | 97.0 | 583 | 91.4 | 767 | 81.2 | 1479 | 85.6 | |
| Total | 133 | 100 | 638 | 100 | 957 | 100 | 1728 | 100 | |
| Route of infection | |||||||||
| IDU | 36 | 30.3 | 134 | 37.0 | 290 | 47.7 | 460 | 42.2 | <0.001 |
| Transfusion-associated | 49 | 41.2 | 117 | 32.3 | 141 | 23.2 | 307 | 28.2 | |
| Nosocomial/parenteral | 16 | 13.4 | 56 | 15.5 | 78 | 12.8 | 172 | 15.8 | |
| Sporadic | 18 | 15.1 | 55 | 15.2 | 99 | 16.3 | 150 | 13.8 | |
| Total | 119 | 100 | 362 | 100 | 608 | 100 | 1089 | 100 | |
| Age | IQR | Age | IQR | Age | IQR | p-Value | |
|---|---|---|---|---|---|---|---|
| Genotype median age | |||||||
| 1 | 35 | 16 | 41 | 18 | 44 | 42 | <0.001 |
| 2 | 41 | 22 | 49 | 12 | 53 | 51 | 0.112 |
| 3 | 29 | 12 | 37 | 10 | 41 | 39 | <0.001 |
| 4 | 27 | 11 | 39 | 10 | 42 | 41 | <0.001 |
| 1a | 29 | 13 | 38 | 12 | 42 | 39 | <0.001 |
| 1b | 37 | 17 | 43 | 20 | 51 | 46 | <0.001 |
IQR: interquartile range; IDU: intravenous drug use.
All study variables, except polymorphism rs12979860, showed multiple significant cross-associations in contingency tables (see supplementary data). We therefore performed binary logistic regression analysis with genotypes 1a, 1b, 2, 3 and 4 as dependent variables, and route of infection, HIV co-infection, immigration status and geographical area of origin as independent variables. All the models were adjusted by age and gender. The marked differences between genotypes 1a and 1b argued against the implementation of a specific model for genotype 1. The categories “Sporadic infection”, “Western Europe” and “1980–1994 period” acted as reference for their respective variables.
Table 3 shows the OR and 95% CIs of the demographic variables significantly associated with HCV genotypes in the multivariate analysis. Neither immigration status nor HIV coinfection was selected in any of the genotype models. Genotype 1b was positively associated with the transfusion-associated and parenteral/nosocomial routes as well as with immigrants from Eastern Europe and showed a negative association with IDU. Genotype 2 was also negatively associated with IDU and positively with immigration from Africa. Genotype 3 had positive associations with IDU, with immigrants from Eastern Europe and especially with immigrants from Asia. Both genotypes 3 and 4 were negatively associated with transfusion-associated and nosocomial/parenteral infections. The last period was positively associated with genotype 3 and 4 and negatively with genotype 1b. The immigrant factor seen in genotype 3 was not shared by genotypes 1a and 4, although these three genotypes did share some other demographic characteristics. There was a significant association of genotype 4 infection with native patients infected via the IDU route (p=0.045) (see supplementary data).
Relationship between selected HCV genotype and demographic variables.
| Genotype | 1a | 1b | 2 | 3 | 4 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | 95% CIs | p–Value | OR | 95% CIs | p–Value | OR | 95% CIs | p–Value | OR | 95% CIs | p–Value | ||
| Route of infection | |||||||||||||
| Sporadic | – | Ref | Ref | Ref | Ref | ||||||||
| IDU | – | 0.343 | 0.203–0.580 | <0.001 | 0.142 | 0.041–0.492 | <0.001 | 1.952 | 1.161–3.284 | 0.009 | – | ||
| Transfusion-associated | – | 2.873 | 1.784–4.626 | <0.001 | – | 0.274 | 0.135–0.556 | <0.001 | 0.413 | 0.182–0.935 | 0.034 | ||
| Parenteral/nosocomial | – | 2.693 | 1.562–4.643 | <0.001 | – | 0.348 | 0.159–0.762 | 0.008 | – | ||||
| Geographical origin | |||||||||||||
| Westen Europe | – | Ref | Ref | Ref | Ref | ||||||||
| Africa | – | – | 22.246 | 4.997–99.040 | <0.001 | – | – | ||||||
| Eastern Europe | – | 2.983 | 1.343–6.627 | 0.007 | – | 2.852 | 1.303–6.244 | 0.009 | – | ||||
| Latin America | – | – | – | – | – | ||||||||
| Asia | – | – | – | 25.370 | 2.915–220.833 | 0.003 | – | ||||||
| Time periods | |||||||||||||
| 1980–1994 | – | Ref | Ref | Ref | Ref | ||||||||
| 1995–2005 | – | – | – | 2.722 | 1.202–6.166 | 0.016 | – | ||||||
| 2006–2015 | – | 0.344 | 0.202–0.588 | <0.001 | – | 3.150 | 1.415–7.014 | 0.005 | 2.504 | 1.131–5.542 | 0.024 | ||
The data presented here reflect the specific rates of HCV genotypes in our geographical area and confirms many of the associated demographic characteristics and temporal trends reported previously.1,5,13,15,16,23–30 In agreement with many studies from Western European countries, the most frequently detected genotype in this report was 1b (42.4%) followed by 1a (22.5%), 3 (18.6%), 4 (10.6%) and 2 (4.6%). This profile is similar to others recently described in Spain,1,25–27 especially in cohorts with a low proportion of HIV-coinfected patients. As expected, the genotypic profile of the 224 HIV-coinfected patients in this series, with higher rates of genotypes 1a, 3 and 4 and lower rates of genotypes 1b and 2, is similar to the ones found in other cohorts of HIV-coinfected patients in Spain,31,32 in whom IDU was also the most prevalent source of infection.
Our study confirms that gender and age are variables closely associated with certain genotype distributions. Genotypes 1b and 2, with median ages above 45, were more prevalent in females than genotypes 1a, 3 and 4, which in turn were more prevalent in males and in patients younger than 45 years.
There were significant associations between certain routes of acquiring the infection and specific genotypes, in accordance with many previous studies. IDU was mainly associated with genotypes 1a, 3 and 4, the transfusion-associated infection with genotypes 1b and 2 and parenteral/nosocomial infections with 1b,1,5,12,13,15,16,22–30,33–36 also confirming the main trends recently reported in our area in the population infected by IDU.37 Logistic regression analysis confirmed positive associations between transfusion and parenteral/nosocomial transmissions and genotype 1b and between IDU and genotype 3 and, conversely, negative associations between genotypes 1b and 2 and IDU and between genotypes 3 and 4 and infections acquired through the transfusion-associated and parenteral/nosocomial routes. As for sporadic infections, in contrast to several studies which reported a genotype distribution similar to that acquired through the transfusion-associated route,24,29,30 we found a relatively homogeneous genotype profile.
Immigrants accounted for 14% of our series, reflecting the high level of recent immigration in the regions of Girona, which rose from 6.2% of the population in the year 2000 to 21.5% in 2010, falling slightly to 19.1% in 2015.38,39 We found a very strong association of genotype 2 with African immigrants, principally sub-Saharans, and of genotype 3 with patients from Asia and Eastern European countries. In almost 20% of the immigrants it was impossible to determine the region of origin, but in this subgroup the distribution of genotypes was very similar to that found in immigrants from Western Europe and Latin America.
Although this series includes patients identified over more than three decades, the information should be considered with a certain amount of caution. First of all, differences in the sensibility and specificity of the techniques used over time could have played a certain role in the prevalence of specific genotypes. Secondly, the possible epidemiologic changes over time may have not necessarily coincided with the periods fixed in the study, as has been pointed out before.31 Finally, there may have been an inherent selection bias due to the retrospective nature of the study. This may have been particularly relevant in the first study period because of the comparatively small sample size and the profile of patients, all of them consecutive outpatients seen at a single office for chronic hepatitis. In the two other periods the larger sample sizes and a diverse origin of patients make a selection bias much more unlikely. However, as the majority of the changes seen in this study also occurred between the two last periods, we considered that a selection bias did not affect the main conclusions. Over these three decades, a decrease in the prevalence of genotype 1b was observed, as well as an increase in the prevalence of genotypes 1a, 3 and 4, as reported extensively in Europe.12,13,24–30,34–36,40 Our study tentatively suggests that the significant increase in the prevalence of genotype 3 might have taken place mainly in the period 1995–2005, while the increasing presence of genotypes 1a and 4 is more recent. The presence of immigrants from certain the geographical areas may have played a role in the increase in genotype 3 and may have maintained its prevalence in the third period. Genotypes 3 and 4 showed no association with immigration but genotype 4 had a specific relationship with native population and the IDU route.
The eradication of transfusion-associated hepatitis and the use of disposable material in health practices, especially in the developed world,41 accounts for the reduction in infections by genotypes 1b and 2, which were linked to these transmission routes. It also explains the trend toward increases in genotypes 1a, 3 and 4, and why IDU remains the most prevalent source of infection in the developed world. Migratory movements toward developed countries might have contributed to the increases in genotypes 3 and 4. However, in Western Europe, a slowdown,40 a decrease32,34 or a stabilization24,35 of genotype 3 prevalence have recently been reported. In our series, the stabilization of the prevalence of genotype 3 was attributable to Asian immigrants infected by this genotype in the period 2006–2015. This change in the prevalence of genotype 3 infection has been attributed to the saturation of the population susceptible to IDU,40 the improvement of risk behaviors among the IDU42 population, and a selection bias of patients in the years previous to the determination of the genotype. Patients who attained SVR with interferon or interferon-ribavirin, much higher in infections with genotypes 2 and 3, would have been excluded from genotyping.34,40 However, the first two hypotheses could also be applied to genotype 1a and 4 infections, also associated with IDU, while the third hypothesis would have involved so few patients in our series that its relevance would be negligible. In addition, immigration contributed very few patients with genotypes 1a and 4, confirming that in our area it is the infection among the native population that explains the increase in genotype 4, mainly linked to the IDU.
Genotype 4 infection in the Spanish native population was reported in 1997,43 especially with the 4c/4d subtype, and less with the 4a subtype26,32,43,44 and mainly in HIV-coinfected patients infected through the IDU route.20,45–48 Our series of 183 mainly native patients with genotype 4 conformed to this pattern, with only four infected individuals from sub-Saharan Africa and a single HIV-positive immigrant, (a hemophiliac from Latin America), among the 35 HIV coinfections with this genotype.
Phylogenetic studies confirm that genotype 4 HCV has an African origin, and was probably introduced in Western Europe through its southern countries,20 in the 1950s48 by the IDU infection route. The infection spread rapidly, first in drug users and then in the general population, and did not stabilize until the 1980s.47 Since the exclusion of paid blood donors was not mandatory until the late 1980s,49 HCV identification was not possible until 198950 and first-generation ELISA was not applied in blood banks until 1991.51 As a result, there was time enough for genotype 4 infection to spread into the general population by mechanisms other than IDU. In fact, 34% of our genotype 4 infected patients reported routes of infection other than IDU, confirming the importance of other transmission routes of genotype 4 infection in our study area.
The increasing prevalence of genotype 1a and 4 infections in our region has coincided with the aging of their respective cohorts. This age factor points toward a stabilization or reduction in the incidence of new infections at younger ages in recent years. Although this is common to all genotypes, it may be especially relevant for genotypes 1a and 4, whose cohorts include a low proportion of immigrants. It is possible that the last cohorts of genotype 4 include infections acquired many years ago, with patients who have recently requested treatment due to the greater social awareness of the disease and the efficacy and applicability of the new oral drugs.
In summary, over the period 1980–2015 we witnessed sustained falls in transfusion-associated and parenteral/nosocomial infection routes and in the prevalence of genotype 1b. These changes have run in parallel with the increasing prevalence of genotypes 1a, 3 and 4, associated with male gender and with IDU, which has become the most frequent route of infection. Immigration has also affected the genotypic profile by contributing genotype 2 infections from Africa and genotype 3 infections from Eastern Europe and Asia. The increasing prevalence of genotypes 1a and 4 appears to be mainly restricted to native patients and to IDU. This study confirms that, although the treatment of chronic hepatitis C should no longer be genotype-dependent, its determination will remain an essential epidemiological tool.
FundingNone declared.
Ethical approvalThis work was approved by the regional ethics committee, CEI GIRONA, under the code name of GENOGIR.
Informed consentIt was considered not necessary due to the retrospective nature of the study and the anonymity of the results.
Conflict of interestThe authors reported no conflict of interests in relation to this study.