Although the pathogenesis of Autoimmune Hepatitis (AIH) is unknown, the susceptibility is determined in part by genes linked to the region of HLA class II. The study included 47 unrelated individuals with the diagnostic of type 1 AIH. A control group (n=81) of healthy individuals was included. The alleles were tested for HLA class II genotyping using polymerase chain reaction and sequence-specific primers technique (PCR-SSP). Among patients with AIH alleles occurring at higher frequencies were DRB1*04, DRB1*03, DRB1*01, DRB1*07 and DRB1*13. With reference to controls the following alleles were the most prevalent DRB1*07, DRB1*11, DRB1*08, DRB1*13 and DRB1*01. In comparison with the control group, AIH patients revealed a greater occurrence of the DRB1*03 and DRB1*04. These alleles can be considered as predisposing factors for the development of AIH. By contrast, DRB1*08 and DRB1*11 alleles were less frequent among patients and hence could be involved in disease resistance.
Aunque la patogenia de la Hepatitis Autoinmune (AIH) es desconocida, la susceptibilidad está determinada en parte por genes HLA de Clase II. En el estudio participaron 47 individuos no relacionados con diagnóstico de AIH tipo 1. Se incluyó un grupo de control (n=81) de individuos sanos. Los alelos HLA de clase II fueron tipificados mediante la reacción en cadena de la polimerasa y la técnica de cebadores de secuencia específica (PCR-SSP). En los pacientes con AIH, los alelos con mayores frecuencias fueron: DRB1*04, DRB1*03, DRB1*01, DRB1*07 y DRB1*13. En los controles los alelos más prevalentes fueron: DRB1*07, DRB1*11, DRB1*08, DRB1*13 y DRB1*01. En comparación con el grupo control, los pacientes con AIH mostraron una mayor incidencia de: DRB1*03 y DRB1*04, que pueden ser considerados como factores predisponentes. En cambio, los alelos DRB1*08 y DRB1*11 de menor frecuencia, estarían relacionados con la resistencia a esta enfermedad.
Autoimmune Hepatitis (AIH) is an inflammation of the liver characterized by the presence of periportal hepatitis in microscopic examination, hypergammaglobulinemia, and serum autoantibodies. Infiltration of the liver by T lymphocytes and a peripheral defect in T suppressor cell function suggest an immune basis for the pathogenesis and liver injury in this disease.1,2
The Major Histocompatibility Complex (MHC) genes, mainly HLA-DR alleles, are implicated in susceptibility and resistance to many diseases, especially autoimmune pathology.3 It is important to consider with different HLA antigens which are associated to disease, according to the ethnic group affected. Therefore, the HLA-disease relationship can vary; the same pathology may be associated with dissimilar HLA alleles. The different associated alleles described in certain populations also reflect dissimilar HLA distributions among populations. Several studies in diverse ethnic group have shown that the genetic susceptibility to AIH is strongly associated with different HLA-DR alleles.4–6 These discrepant observations emphasize the importance of studying different populations.
In the present study, we performed HLA genotyping in Argentinean patients to type 1 AIH using PCR amplification by the sequence-specific primers (PCR-SSP) method to investigate the contribution of the DRB1 highly polymorphic locus in determining the susceptibility to the disease in subjects from Rosario, Santa Fe Province, Argentina. Urban populations of Argentina are assumed to have a white Caucasian European genetic component. However, historical and biological data account for the influence of other ethnic groups.7–9 Therefore, the present study allows analyzing the genetic association in a sample of this population.
Subjects and methodsWe studied 47 samples of peripheral blood obtained by venipuncture from patients to type 1 AIH who attended medical advice and control to the Gastroenterology Service, Hospital Provincial del Centenario. Patients included in this study did not have other liver disease. This group consists of 40 females and 7 males, average age of 26.9±11.9 years. Cases were from Rosario city, which is the third largest urban area in the country.
Additionally, 81 non-related healthy individuals with neither symptoms nor previous diagnosis of AIH were included as a control group. Those individuals were ethnically matched patients from the same geographic area.
All subjects gave informed consent to participate in the study, and the protocol was approved by the Ethic Committee of the School of Medical Sciences of Rosario, Argentina, according to the principles of the Declaration of Helsinki.
DNA was obtained from EDTA anticoagulant peripheral blood by the method of CTAB (hexadecyltrimethylammonium bromide).10
Washing the blood sample with TE (Tris EDTA) buffer was made in order to eliminate the hemoglobin of red blood cells, and Fe that may interfere later in the PCR amplification reactions. During incubation with CTAB, cell membranes were lysed and the DNA passes into the suspension. The proteins were extracted with chloroform. The addition of isopropanol precipitated DNA from the aqueous fraction. Washing with 70% ethanol was performed to remove the isopropanol.
The alleles were tested using polymerase chain reaction and sequence-specific primers technique (PCR-SSP), and the typing technique used is of low resolution.11
Each PCR reaction mix contained alleles or group-specific DRB1 primers and the internal positive control primer pair in a lower concentration. These two primers matched non-allelic sequences and thus functioned as an internal positive amplification control. 5′-primer and 3′-primer gave rise to a 796 base pair (bp) fragment. Each PCR reaction mixture contained 2–4 allele or group-specific DRB1 primers and the internal positive control primer pair. The optimum conditions for amplification were established by trial and error to obtain enough highly specific products. The PCR products were evaluated by gel electrophoresis 2% agarose and stained with ethidium bromide. For each individual, the alleles were assigned according to the presence or absence of amplification.
Differences in the distribution of DRB1 alleles between AIH disease patients and controls were analyzed by or Pearson's x2 test or Fisher's exact test, if applicable. Odds Ratios (OR) was calculated. P-values<0.05 were considered significant and further corrected according to the number of comparisons.12
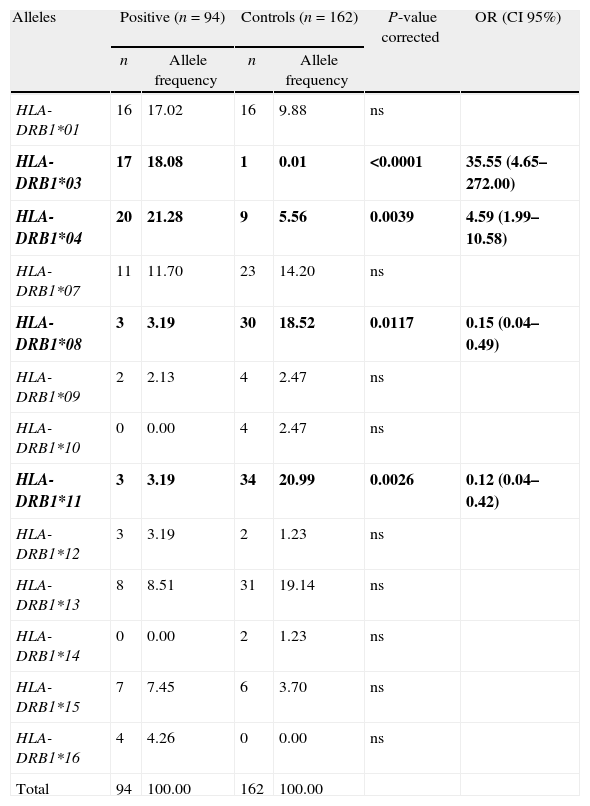
Results and discussionThe distributions of the DRB1 alleles in patients and controls are summarized in Table 1. Individuals with AIH and healthy controls revealed 11 and 12 DRB1 different alleles, respectively. Among patients with AIH alleles occurring at higher frequencies (%) were DRB1*04 (21.28%), DRB1*03 (18.08%), DRB1*01 (17.02%), DRB1*07 (11.70%) and DRB1*13 (8.51%). With reference to controls the following alleles were the most prevalent: DRB1*11 (20.99%), DRB1*13 (19.14%), DRB1*08 (18.52%), DRB1*07 (14.20%) and DRB1*01 (9.88%).
Between-group comparisons of patients with type 1 AIH and controls.
| Alleles | Positive (n=94) | Controls (n=162) | P-value corrected | OR (CI 95%) | ||
| n | Allele frequency | n | Allele frequency | |||
| HLA-DRB1*01 | 16 | 17.02 | 16 | 9.88 | ns | |
| HLA-DRB1*03 | 17 | 18.08 | 1 | 0.01 | <0.0001 | 35.55 (4.65–272.00) |
| HLA-DRB1*04 | 20 | 21.28 | 9 | 5.56 | 0.0039 | 4.59 (1.99–10.58) |
| HLA-DRB1*07 | 11 | 11.70 | 23 | 14.20 | ns | |
| HLA-DRB1*08 | 3 | 3.19 | 30 | 18.52 | 0.0117 | 0.15 (0.04–0.49) |
| HLA-DRB1*09 | 2 | 2.13 | 4 | 2.47 | ns | |
| HLA-DRB1*10 | 0 | 0.00 | 4 | 2.47 | ns | |
| HLA-DRB1*11 | 3 | 3.19 | 34 | 20.99 | 0.0026 | 0.12 (0.04–0.42) |
| HLA-DRB1*12 | 3 | 3.19 | 2 | 1.23 | ns | |
| HLA-DRB1*13 | 8 | 8.51 | 31 | 19.14 | ns | |
| HLA-DRB1*14 | 0 | 0.00 | 2 | 1.23 | ns | |
| HLA-DRB1*15 | 7 | 7.45 | 6 | 3.70 | ns | |
| HLA-DRB1*16 | 4 | 4.26 | 0 | 0.00 | ns | |
| Total | 94 | 100.00 | 162 | 100.00 | ||
OR, odds ratio; CI, confidence interval; ns, not significant.
Data represent the number of each allele detected (n) and the allele frequency with respect to the total number of studied alleles. P-values<0.05 were considered significant and further corrected according to the number of comparisons.
Bold, indicates the alleles present OR with significant differences between both groups.
In comparison with the control group, AIH patients revealed a greater occurrence of the DRB1*03 (P<0.0001; OR: 35.55) and DRB1*04 (P=0.0039; OR: 4.59), for which they may be implicated in AIH. By contrast, DRB1*08 (P=0.0117; OR: 0.15) and DRB1*11 (P=0.0026; OR: 0.12) alleles were less frequent among patients and could be involved in disease resistance.
It was considered that some HLA alleles might contribute to the development of AIH. Therefore, in this work we studied in a group of patients the city of Rosario, Argentina, the association the DRB1 alleles with type 1 AIH. Molecular typing allowed the identification of increased frequencies of DRB1*03 and DRB1*04, suggesting that these alleles could be related with this hepatic pathology. Another interesting finding was that DRB1*08 and DRB1*11 could be associated with disease resistance. These findings should be studied by molecular techniques that allow us to higher resolution of alleles identify you subtypes found.
Various authors have found associations between certain HLA alleles and AIH, in different ethnic groups. DNA-based techniques indicate that DRB1*0301 is the principal risk factor for this disease among Caucasian, and DRB1*0401 had secondary predisposing effect in the same population.4 Tanwandee and associates in Thailand found that the DRB1*0301 was significantly associated with the development of AIH.5 In Latin America, Duarte Rey et al. established positive association with DRB1*0405 and DRB1*1301 alleles while the DRB1*1302 showed a negative association with the disease.6 Fainboim et al. found in Argentinean pediatric patients susceptibility with DRB1*1301, DRB1*0301 and DRB1*1302.13,14 More studies are needed to validate this result in different populations.
Importantly, the prevalence of DRB1*03 and DRB1*04 observed in our patients is in concordance with previous studies reported that these alleles are associated with AIH in North Americans, Western Asians and in North European descendents.4–6 In addition, we observed that the allele frequencies of control samples are not concordant with published data, indicating the contribution of alleles of other ethnicity. Urban populations of Argentina are assumed to have a predominantly white Caucasian European genetic component as a consequence of the massive immigration at the beginning of the 20th century.7 However, recent biological information revealed that Argentina's population of some cities should be considered as a hybrid of Europeans, Amerindians and Africans.8,9,15 Our findings contribute to question the assumptions of a mainly European origin and reveal a more multi-ethnic identity of the population in the Rosario region of central Argentina.
Knowing the influence of the HLA system in the genetic susceptibility to the development of type 1 AIH, allow a better understanding of its pathogenic mechanism. However, we must state that the HLA genotype alone does not determine the possibility of developing a disease. The presence or absence of an MHC allele is not an absolute relation to pathology. These associations are not strictly causal relationships, as conferred by the HLA allele variants, not the disease itself but the tendency or predisposition to develop it. Similarly, it is inaccurate to award an absolute value to the presence of the protective alleles.
Conflict of interestThe authors declare that they have no conflict of interest.