The aim of this study was to estimate the diversity and prevalence of both groups of Brucella canis 1 and 2 with and without deletion respectively in different areas of Argentina. A total of 104 bacterial cultures were typed as B. canis strains using the classical biotyping method. Two PCR assays were performed to confirm that all isolates were B. canis and not Brucella suis. The differentiation between groups 1 and 2 was achieved using another PCR assay and the diversity of B. canis isolates was assessed with four MLVA_16 markers. All strains belonged to Group 2. Bruce 09 marker (MLVA_16 assay) showed the greatest diversity. Only Group 2 of B. canis was identified among the strains evaluated. The markers chosen from the MLVA_16 allowed us to detect genetic diversity among the strains of B. canis studied.
El objetivo de este trabajo fue estimar la diversidad y la prevalencia de ambos grupos de Brucella canis 1 y 2 (con y sin deleción, respectivamente) en diferentes áreas de Argentina. Un total de 104 cultivos bacterianos se tipificaron como cepas de B. canis usando biotipado clásico. Se realizaron dos ensayos de PCR para confirmar que todos los aislamientos eran B. canis y no Brucella suis. La diferenciación entre los grupos 1 y 2 se logró con otro ensayo de PCR, y la diversidad entre las cepas de B. canis se obtuvo mediante el empleo de cuatro marcadores del ensayo de MLVA_16. Todas las cepas pertenecieron al grupo 2. El marcador Bruce 09 (ensayo MVLA-16) mostró la mayor diversidad. Sólo se halló el Grupo 2 de B. canis entre las cepas estudiadas. Los marcadores del MLVA_16 permitieron detectar la presencia de diversidad genética entre las cepas de B. canis analizadas.
Brucella canis is an etiological agent of canine brucellosis that has been found in many countries, such as the US, China and Mexico as well as in countries of South America, Asia and Europe. This zoonotic disease causes abortion in female dogs and epididymitis in male dogs as the main symptom. In addition, this disease causes important losses in breeding kennels because of reproductive failures5.
Bacterial isolation is the “gold standard” for the definitive diagnosis, despite the low sensitivity of this procedure regarding clinical samples2. In addition, the global scientific community uses the process of phenotypic characterization from isolates to distinguish the different species of the genus Brucella1.
Researchers have employed the polymerase chain reaction assay (PCR) in many studies for the molecular typing of Brucella species and biovars from bacteriological cultures10,14. For example, the Bruce-ladder assay (a multiplex PCR assay) is the only analysis accepted by the World Organization for Animal Health (OIE) for identification and typing Brucella species13. However, some B. canis strains can be identified erroneously as B. suis10. The researchers that reported this finding identified two groups of B. canis based on the presence or absence of a region of the BMEI1435 polysaccharide deacetylase gene in B. canis. Indeed, Group 1 presents a deletion (PCR amplifies a 607-bp fragment), whereas Group 2 lacks the deletion (PCR amplifies a 1674-bp fragment). López Goñi et al. replaced two primers of the PCR original protocol and obtained the Bruce-ladder v2.0 test14. This new test allows the differentiation of strains of B. canis, five biovars of B. suis and B. neotomae. In parallel, Kang et al.9 also developed new primers from a specific region to avoid the problem of the original Bruce-ladder.
Another research group found that only Group 2 is present in Medellín city15. More recently, another study reported both groups in China4.
Multiple Locus Variable-number Tandem Repeat Analysis (MLVA) is a method that has been performed for the typing of bacterial species including Mycobacterium tuberculosis, Bacillus anthracis and others. A set of eight microsatellite loci has been proposed because they are highly discriminatory and efficient for distinguishing strains within a local outbreak. Although we cannot correctly predict the biovar or even the species of an isolate, this method is a useful tool especially in outbreaks. To date, different MLVA assays have been developed for typing Brucella strains with different kind of outcomes and utilities12.
To the best of our knowledge, in Argentina, there are no studies published on groups of B. canis isolates or any publication on B. canis MLVA studies. With this in mind, the aim of this study was to estimate the prevalence of both groups of B. canis in different areas of Argentina from microbiological isolates of clinical dog samples. Other objectives were to compare the performance of two PCR assays in discriminating B. canis and B. suis strains and, finally, to estimate the diversity of B. canis using four markers of the MLVA_16 assay.
The analyzed samples consisted of 104 B. canis isolates from clinical dog samples obtained between August 2009 and December 2018. The samples were collected from the Autonomous City of Buenos Aires and different areas of Argentina. Strains were typed following the recommendations described elsewhere1. Molecular typing confirmation of all B. canis strains and not B. suis was performed using two PCR assays9,14. DNA was extracted using a commercial kit, ROCHE High Pure PCR Template Preparation Kit (Roche Diagnostics, GmbH Roche Applied Science, Germany), following the manufacturer's instructions. A concentration between 50 and 70 ng/μl was used for further analyses after measuring DNA with ND-1000 NanoDrop. As a negative control of DNA extraction, we used ultrapure water. Two PCR assays (PCR1 and PCR2) were carried out to differentiate B. canis and B. suis. PCR1 and PCR2 were performed according to Lopez Goñi et al.14 and Kang et al.9 respectively. For both PCRs, B. canis strain RM6/66 and B. suis biovar 1 were used as positive controls. In addition, we used ultrapure water as a negative control. A third PCR (PCR3) was conducted according to Koylass et al.10B. canis strain RM6/66 (Group 1) and Brucella melitensis biovar 1 (Group 2) were used as positive controls, respectively. All the products from PCR1, PCR2 and PCR3 were analyzed by electrophoresis in 1.5% agarose gel stained with ethidium bromide (0.5μg/ml) and UV light visualization.
The markers used for the MLVA assay in this study were Bruce 04, Bruce 07, Bruce 09 and Bruce 1612. The selection of these markers was based on their greatest diversity among species of B. canis4,8 (Table S1). A total of 15 strains were selected from various areas of Argentina: San Miguel de Tucumán (Province of Tucumán); Rio Cuarto (Province of Córdoba); Rosario (Province of Santa Fe); Río Grande (Province of Tierra del Fuego); General Pico (Province of La Pampa); Corrientes (Province of Corrientes); Ituzaingó, Tortuguitas, Paso del Rey, Del Viso (two strains), Villa Ballester, Banfield, Verónica (Province of Buenos Aires) and Villa Lugano (Autonomous City of Buenos Aires). This allowed an approximate representation of the different alleles (number of repeats) present in different areas of the country.
DNA amplification was conducted in tubes with a final volume of 25μl per reaction. Each reaction contained DNA (50μg), each primer (forward/reverse; 0.5μM), each of the four deoxynucleotide triphosphates (dNTPs; 200μM), 1X buffer with magnesium chloride (MgCl2, final concentration of 1.5mM (PROMEGA 5X Green GoTaq® Flexi Buffer Migration Pattern) and 1 U of Taq DNA Polymerase. B. canis strain RM6/66 and B. melitensis 16M were used as positive controls. Ultrapure water was used as negative control. The thermocycling conditions were as follows: a first denaturation at 94°C for 3min and 30 cycles of 94°C for 30s, 60°C for 30s and 72°C for 50s with a final extension of 72°C for 5min. Bionumerics software version 3.5 (Applied Maths, St-Martens-Latem, Belgium) was used to estimate the size of the PCR bands of the agarose gels. The quantification of the number of repeats of each marker was performed following the protocol described by Le Flèche et al.12 The dendrogram was generated through the unweighted pair group method following arithmetic averages (UPGMA) and using the PAST software6. The Hunter-Gaston Diversity Index (HGDI)7 was calculated using Epicompare software version 1.0 (www.ridom.de/epicompare) to elucidate the discriminatory power of the genotyping methods. Number 0 and 1 stand for “without diversity” and “extreme diversity”, respectively. These results reflect the total of detected alleles. The PCR products were analyzed in 2% agarose gel in Tris acetate and EDTA (ethylenediaminetetraacetic acid) 1X buffer (TAE) with the addition of 0.5μg/ml of ethidium bromide. An Applied Biosystems Veriti™ Thermal Cycler was used for the PCR reactions.
All 104 strain isolates were typed as B. canis according to the classical biotyping method. In this study, we evaluated two PCR assays for molecular typing of the evaluated bacteria that had been confirmed as B. canis strains by the classical method.
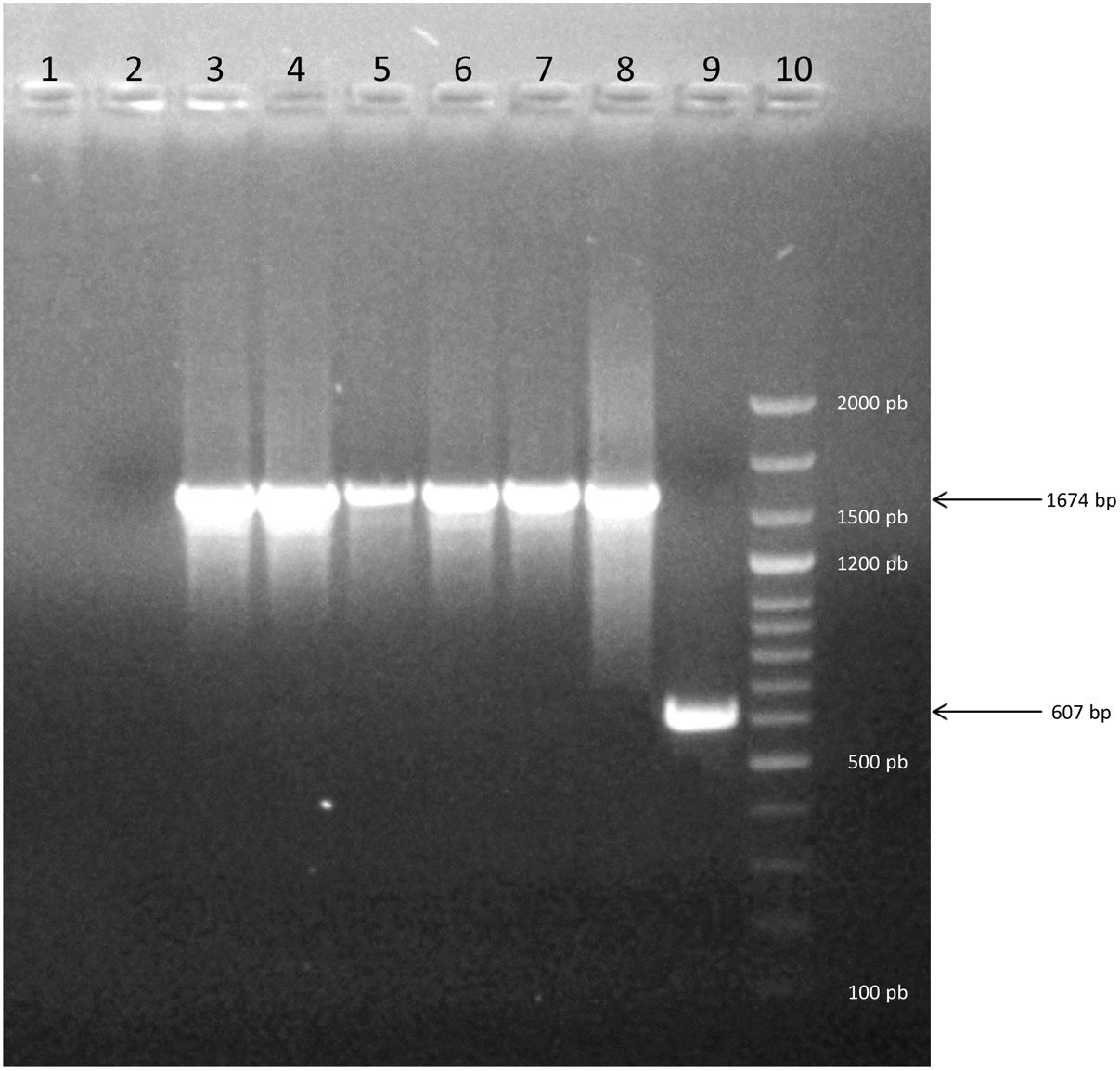
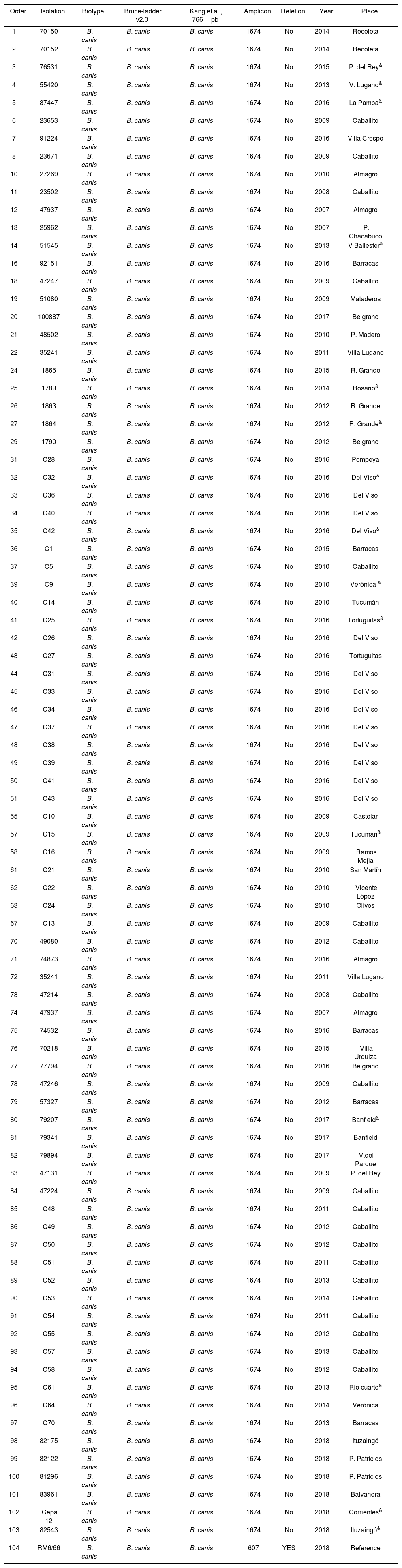
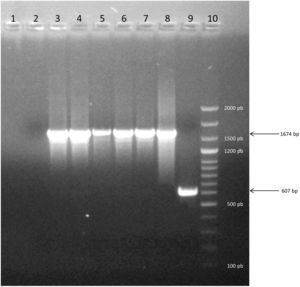
The two PCRs used to assess the species (PCR1 and PCR2) confirmed that all strains were B. canis (Table 1). Thus, both techniques yielded consistent results. PCR1 shows the molecular patterns between strains of B. canis and B. suis. PCR2 displays a 776-bp differential amplicon between species, i.e. B. suis contains the amplicon, whereas B. canis lacks it. In this study, all the evaluated strains were B. canis Group 2 (without deletion; 100% (CI 95% 0.95–1.00); Table 1, Fig. 1).
Description of B. canis strains and techniques used for their characterization and differentiation in groups 1 and 2.
| Order | Isolation | Biotype | Bruce-ladder v2.0 | Kang et al., 766pb | Amplicon | Deletion | Year | Place |
|---|---|---|---|---|---|---|---|---|
| 1 | 70150 | B. canis | B. canis | B. canis | 1674 | No | 2014 | Recoleta |
| 2 | 70152 | B. canis | B. canis | B. canis | 1674 | No | 2014 | Recoleta |
| 3 | 76531 | B. canis | B. canis | B. canis | 1674 | No | 2015 | P. del Rey& |
| 4 | 55420 | B. canis | B. canis | B. canis | 1674 | No | 2013 | V. Lugano& |
| 5 | 87447 | B. canis | B. canis | B. canis | 1674 | No | 2016 | La Pampa& |
| 6 | 23653 | B. canis | B. canis | B. canis | 1674 | No | 2009 | Caballito |
| 7 | 91224 | B. canis | B. canis | B. canis | 1674 | No | 2016 | Villa Crespo |
| 8 | 23671 | B. canis | B. canis | B. canis | 1674 | No | 2009 | Caballito |
| 10 | 27269 | B. canis | B. canis | B. canis | 1674 | No | 2010 | Almagro |
| 11 | 23502 | B. canis | B. canis | B. canis | 1674 | No | 2008 | Caballito |
| 12 | 47937 | B. canis | B. canis | B. canis | 1674 | No | 2007 | Almagro |
| 13 | 25962 | B. canis | B. canis | B. canis | 1674 | No | 2007 | P. Chacabuco |
| 14 | 51545 | B. canis | B. canis | B. canis | 1674 | No | 2013 | V Ballester& |
| 16 | 92151 | B. canis | B. canis | B. canis | 1674 | No | 2016 | Barracas |
| 18 | 47247 | B. canis | B. canis | B. canis | 1674 | No | 2009 | Caballito |
| 19 | 51080 | B. canis | B. canis | B. canis | 1674 | No | 2009 | Mataderos |
| 20 | 100887 | B. canis | B. canis | B. canis | 1674 | No | 2017 | Belgrano |
| 21 | 48502 | B. canis | B. canis | B. canis | 1674 | No | 2010 | P. Madero |
| 22 | 35241 | B. canis | B. canis | B. canis | 1674 | No | 2011 | Villa Lugano |
| 24 | 1865 | B. canis | B. canis | B. canis | 1674 | No | 2015 | R. Grande |
| 25 | 1789 | B. canis | B. canis | B. canis | 1674 | No | 2014 | Rosario& |
| 26 | 1863 | B. canis | B. canis | B. canis | 1674 | No | 2012 | R. Grande |
| 27 | 1864 | B. canis | B. canis | B. canis | 1674 | No | 2012 | R. Grande& |
| 29 | 1790 | B. canis | B. canis | B. canis | 1674 | No | 2012 | Belgrano |
| 31 | C28 | B. canis | B. canis | B. canis | 1674 | No | 2016 | Pompeya |
| 32 | C32 | B. canis | B. canis | B. canis | 1674 | No | 2016 | Del Viso& |
| 33 | C36 | B. canis | B. canis | B. canis | 1674 | No | 2016 | Del Viso |
| 34 | C40 | B. canis | B. canis | B. canis | 1674 | No | 2016 | Del Viso |
| 35 | C42 | B. canis | B. canis | B. canis | 1674 | No | 2016 | Del Viso& |
| 36 | C1 | B. canis | B. canis | B. canis | 1674 | No | 2015 | Barracas |
| 37 | C5 | B. canis | B. canis | B. canis | 1674 | No | 2010 | Caballito |
| 39 | C9 | B. canis | B. canis | B. canis | 1674 | No | 2010 | Verónica & |
| 40 | C14 | B. canis | B. canis | B. canis | 1674 | No | 2010 | Tucumán |
| 41 | C25 | B. canis | B. canis | B. canis | 1674 | No | 2016 | Tortuguitas& |
| 42 | C26 | B. canis | B. canis | B. canis | 1674 | No | 2016 | Del Viso |
| 43 | C27 | B. canis | B. canis | B. canis | 1674 | No | 2016 | Tortuguitas |
| 44 | C31 | B. canis | B. canis | B. canis | 1674 | No | 2016 | Del Viso |
| 45 | C33 | B. canis | B. canis | B. canis | 1674 | No | 2016 | Del Viso |
| 46 | C34 | B. canis | B. canis | B. canis | 1674 | No | 2016 | Del Viso |
| 47 | C37 | B. canis | B. canis | B. canis | 1674 | No | 2016 | Del Viso |
| 48 | C38 | B. canis | B. canis | B. canis | 1674 | No | 2016 | Del Viso |
| 49 | C39 | B. canis | B. canis | B. canis | 1674 | No | 2016 | Del Viso |
| 50 | C41 | B. canis | B. canis | B. canis | 1674 | No | 2016 | Del Viso |
| 51 | C43 | B. canis | B. canis | B. canis | 1674 | No | 2016 | Del Viso |
| 55 | C10 | B. canis | B. canis | B. canis | 1674 | No | 2009 | Castelar |
| 57 | C15 | B. canis | B. canis | B. canis | 1674 | No | 2009 | Tucumán& |
| 58 | C16 | B. canis | B. canis | B. canis | 1674 | No | 2009 | Ramos Mejía |
| 61 | C21 | B. canis | B. canis | B. canis | 1674 | No | 2010 | San Martín |
| 62 | C22 | B. canis | B. canis | B. canis | 1674 | No | 2010 | Vicente López |
| 63 | C24 | B. canis | B. canis | B. canis | 1674 | No | 2010 | Olivos |
| 67 | C13 | B. canis | B. canis | B. canis | 1674 | No | 2009 | Caballito |
| 70 | 49080 | B. canis | B. canis | B. canis | 1674 | No | 2012 | Caballito |
| 71 | 74873 | B. canis | B. canis | B. canis | 1674 | No | 2016 | Almagro |
| 72 | 35241 | B. canis | B. canis | B. canis | 1674 | No | 2011 | Villa Lugano |
| 73 | 47214 | B. canis | B. canis | B. canis | 1674 | No | 2008 | Caballito |
| 74 | 47937 | B. canis | B. canis | B. canis | 1674 | No | 2007 | Almagro |
| 75 | 74532 | B. canis | B. canis | B. canis | 1674 | No | 2016 | Barracas |
| 76 | 70218 | B. canis | B. canis | B. canis | 1674 | No | 2015 | Villa Urquiza |
| 77 | 77794 | B. canis | B. canis | B. canis | 1674 | No | 2016 | Belgrano |
| 78 | 47246 | B. canis | B. canis | B. canis | 1674 | No | 2009 | Caballito |
| 79 | 57327 | B. canis | B. canis | B. canis | 1674 | No | 2012 | Barracas |
| 80 | 79207 | B. canis | B. canis | B. canis | 1674 | No | 2017 | Banfield& |
| 81 | 79341 | B. canis | B. canis | B. canis | 1674 | No | 2017 | Banfield |
| 82 | 79894 | B. canis | B. canis | B. canis | 1674 | No | 2017 | V.del Parque |
| 83 | 47131 | B. canis | B. canis | B. canis | 1674 | No | 2009 | P. del Rey |
| 84 | 47224 | B. canis | B. canis | B. canis | 1674 | No | 2009 | Caballito |
| 85 | C48 | B. canis | B. canis | B. canis | 1674 | No | 2011 | Caballito |
| 86 | C49 | B. canis | B. canis | B. canis | 1674 | No | 2012 | Caballito |
| 87 | C50 | B. canis | B. canis | B. canis | 1674 | No | 2012 | Caballito |
| 88 | C51 | B. canis | B. canis | B. canis | 1674 | No | 2011 | Caballito |
| 89 | C52 | B. canis | B. canis | B. canis | 1674 | No | 2013 | Caballito |
| 90 | C53 | B. canis | B. canis | B. canis | 1674 | No | 2014 | Caballito |
| 91 | C54 | B. canis | B. canis | B. canis | 1674 | No | 2011 | Caballito |
| 92 | C55 | B. canis | B. canis | B. canis | 1674 | No | 2012 | Caballito |
| 93 | C57 | B. canis | B. canis | B. canis | 1674 | No | 2013 | Caballito |
| 94 | C58 | B. canis | B. canis | B. canis | 1674 | No | 2012 | Caballito |
| 95 | C61 | B. canis | B. canis | B. canis | 1674 | No | 2013 | Rio cuarto& |
| 96 | C64 | B. canis | B. canis | B. canis | 1674 | No | 2014 | Verónica |
| 97 | C70 | B. canis | B. canis | B. canis | 1674 | No | 2013 | Barracas |
| 98 | 82175 | B. canis | B. canis | B. canis | 1674 | No | 2018 | Ituzaingó |
| 99 | 82122 | B. canis | B. canis | B. canis | 1674 | No | 2018 | P. Patricios |
| 100 | 81296 | B. canis | B. canis | B. canis | 1674 | No | 2018 | P. Patricios |
| 101 | 83961 | B. canis | B. canis | B. canis | 1674 | No | 2018 | Balvanera |
| 102 | Cepa 12 | B. canis | B. canis | B. canis | 1674 | No | 2018 | Corrientes& |
| 103 | 82543 | B. canis | B. canis | B. canis | 1674 | No | 2018 | Ituzaingó& |
| 104 | RM6/66 | B. canis | B. canis | B. canis | 607 | YES | 2018 | Reference |
&: strains analyzed by MLVA.
Electrophoresis on agarose gel of PCR products showing representative strains of Group 2. Lane 1: Negative control of PCR. Lane 2: Negative control of DNA extraction. Lane 3–7: profile of five strains evaluated. Lane 8: positive control of Group 2 (1674bp). Line 9: positive control of Group 1 (607bp). Line 10: molecular marker Dangsheng Biotech 100bp DNA ladder plus.
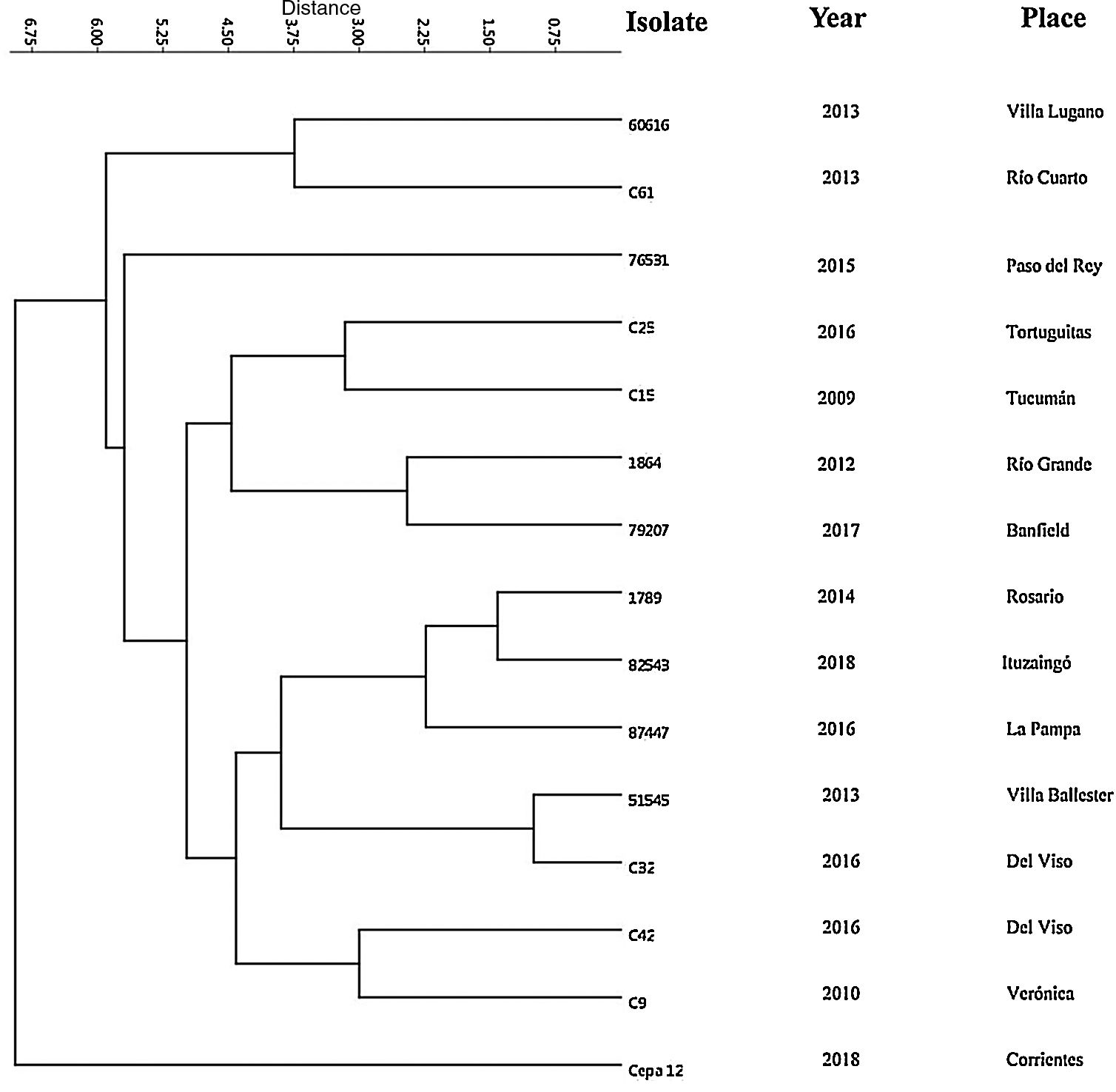
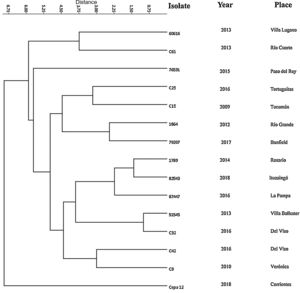
Subsequently, we performed an evaluation of the strains by MLVA_16 by using four markers. For this purpose, we analyzed 15 strains from different regions of Argentina. The overall Diversity Index (HDGI) of the technique was 1.0 (CI 95% 1.0–1.0). The HDGI coefficient ranged from 0.705 to 0.895. Bruce 09 marker displayed the greatest diversity (0.895 CI 95% 0.816–0.974), followed by Bruce 16 (0.876 CI 95% 0.803–0.949), Bruce 07 (0.848 CI 95% 0.805–0.890) and Bruce 04 (0.705 CI 95% 0.550–0.859) (Table S2). Additionally, we identified 15 different molecular patterns (Bruce 04, Bruce 07, Bruce 09, Bruce16 respectively: 4-8-6-9, 6-5-8-5, 6-5-11-8, 6-5-11-9, 6-5-6-5, 6-6-8-9, 6-8-10-11, 6-9-8-8, 7-7-10-5, 8-6-7-9, 8-6-8-8, 8-8-4-12, 8-9-9-11, 8-9-5-5, 11-7-10-7) (Table S3). The molecular pattern detected in Del Viso (6-5-8-5) and the ones found in Banfield (6-8-10-11) were similar to those described in the city of Beijing and the Autonomous Region of Guanxi, Republic of China, respectively4. In addition, the molecular pattern found in La Pampa (6-6-8-9) was similar to those in Paju city, Gyeonggi Province, South Korea8.
In this study, we evaluated various B. canis strains from different regions of Argentina to assess the groups of strains existing in Argentina. All strains were identified using the classical biotyping method and molecular techniques. Among all the strains analyzed in this study, we only detected B. canis type 2 (without deletion of the BMEI1435 gene). This finding is consistent with a previous report from Colombia15. Similarly, most of the isolates analyzed in China belong to this type, although three isolates exhibited a deletion4. By contrast, Koylass et al., found both groups circulating in equal proportion in samples evaluated from Europe, South America and the United States. Specifically, the strains with the deletion came from Peru, Germany and the United States10. The pathogenicity of the strains with and without this deletion has not been studied yet. These data could trigger future research regarding the characteristics of the BMEI1435 gene. This gene has a hydrolase function and participates in the metabolism of carbohydrates. The variation of this gene may have an impact on virulence in the host, but this is still unclear. A recent study compared different Brucella genes and their virulence but did not include BMEI1435 in the analysis, perhaps due to the lack of consideration regarding pathogenicity at the date of the cited study3.
In the present study, we used four markers presented at panel 2 of the MLVA_16 (Bruce 04, Bruce 07, Bruce 09 and Bruce 16) and based on the polymorphism found by other researchers4,8. We did not use the complete panel of MLVA_16 and therefore we believe we cannot speak of “genotypes”. Therefore, we defined them as molecular patterns instead. As a preliminary study, we used 15 strains from different regions of the country, which provided data on the diversity among circulating B. canis strains in specific areas of Argentina. It should be noted that the remaining markers of the full panel of the MLVA_16 in the research of Di et al.4 and Kang et al.8 showed similar repeat numbers. These data suggest high homology of B. canis for these gene regions. Thus, the four markers used in the present study are suitable for evaluating B. canis genetic diversity. However, the use of the complete panel is very useful for other Brucella species such as B. melitensis, B. suis and B. abortus11.
The results from the dendrogram (Fig. 2) suggest great similarity between the strains of Villa Ballester and Del Viso (Province of Buenos Aires) and this result could be explained by the closeness of the locations (36km). Although other strains, such as the strains of the city of Rosario (Province of Santa Fe) and Ituzaingó (Province of Buenos Aires) are somewhat further away (300km), they also showed high similarity. In addition, the similarity between the strains of Rio Grande (Province of Tierra del Fuego) and the town of Banfield (Province of Buenos Aires), which are separated by 2800km, is of particular interest. These findings suggest that dogs may have circulated from one location to another. This is quite usual as people often migrate with their dogs.
Finally, as a first preliminary study in Argentina, our results indicate the existence of a genetic diversity among circulating B. canis strains. In addition, the development of the complete MLVA_16 panel should be done in the near future in order to broaden the knowledge of the circulating genotypes in Argentina.
Our results suggest that the only group of B. canis circulating in Argentina is Group 2 (without BMEI1435 gene deletion). The two PCRs used for molecular typing yielded consistent results. Therefore, both are useful to discriminate between B. canis and B. suis. However, PCR1 should be used to evaluate biovars of B. suis. The use of the four markers chosen from the MLVA_16 allowed us to identify the genetic diversity among the strains of B. canis circulating in Argentina. Thus, as a preliminary test, MLVA_16 is useful for the genetic discrimination of this bacterium.
Conflict of interestThe authors declare that they have no conflicts of interest.
We thank Dr. Gabriela Escobar for the provision of strains, Dr. Zumarraga Martín for processing agarose geles and Dr. Julia Sabio y García for critical reading of the manuscript. This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.