Lithium (Li) is widely distributed in nature and has several industrial applications. The largest reserves of Li (over 85%) are in the so-called “triangle of lithium” that includes the Salar de Atacama in Chile, Salar de Uyuni in Bolivia and Salar del Hombre Muerto in Argentina. Recently, the use of microorganisms in metal recovery such as copper has increased; however, there is little information about the recovery of lithium. The strain Rhodococcus sp. A5wh used in this work was previously isolated from Laguna Azul. The assays revealed that this strain was able to accumulate Li (39.52% of Li/g microbial cells in 180min) and that it was able to grow in its presence up to 1M. In order to understand the mechanisms implicated in Li tolerance, a proteomic approach was conducted. Comparative proteomic analyses of strain A5wh exposed and unexposed to Li reveal that 17 spots were differentially expressed. The identification of proteins was performed by MALDI-TOF/MS, and the obtained results showed that proteins involved in stress response, transcription, translations, and metabolism were expressed under Li stress. This knowledge constitutes the first proteomic approach to elucidate the strategy followed by Rhodococcus to adapt to Li.
El litio (Li) es un elemento químico con múltiples aplicaciones industriales. Es considerado uno de los minerales más ampliamente distribuidos en la naturaleza. Sus mayores reservas (más del 85%) se encuentran en el llamado «triángulo de litio»: salar de Atacama, en Chile; salar de Uyuni, en Bolivia, y salar del Hombre Muerto, en Argentina. En los últimos años, el empleo de microorganismos en la recuperación de metales se ha visto incrementado; sin embargo, hay muy poca información sobre la recuperación de Li por esta vía. En este estudio se trabajó con Rhodococcus sp. A5wh, cepa aislada de Laguna Azul. Los ensayos revelaron que este microorganismo fue capaz de acumular Li (39,52% de Li/g de biomasa en 180 min) y de crecer en presencia de este metaloide hasta una concentración de 1M. Para comprender los mecanismos implicados en la tolerancia al Li, se llevó a cabo el análisis proteómico comparativo de esta cepa expuesta o no expuesta al Li. Los resultados revelaron 17 spots expresados en forma diferencial. La identificación de las proteínas se realizó por MALDI-TOF/MS. Este estudio constituye el primer enfoque proteómico para dilucidar la estrategia seguida por Rhodococcus en su adaptación al estrés.
Lithium is widely distributed on Earth; seawaters contain a concentration of 0.1–0.2 part per million (ppm), whereas in earth, the concentration has been estimated at 65ppm. The largest reserves of Li (over 85%) are found in South America specifically Chile, Argentina and Bolivia, which is known as the “Lithium Triangle” and includes the Salar de Atacama (Chile), Salar de Uyuni (Bolivia), Salar del Hombre Muerto and Salar del Rincón (Argentina). Argentina is the third world producer of Li, associated with the Li reserves of the Puna salt lakes22. Li has many important and interesting uses as heat-resistant ceramics, batteries, pharmaceutical products and it is also required for military, automotive, aircraft, and marine applications. In the last years, the demand for this mineral has increased exponentially1,34,53,66,72,75; therefore the recovery is an important subject of study.
The recovery of Li usually involves solar pond evaporation of the brine. Water evaporates and it does not necessarily return as precipitation to the site of origin, because these ponds are in a high desert environment1,31. The Puna is a desert, which is high in altitude and with the largest thermal amplitude of the world3. The availability of surface water sustains a particular biodiversity and the oldest native people.
An ecologically acceptable and economic alternative is the extraction of metals by means of microorganisms36, through a process known as “biomining or bioleaching”12,13,40,59,60,65,67,74. Microorganisms are being used for the recovery of metals such as copper44,51,52,57,61, uranium32, thorium77 and gold28. However, there is little information on the recovery of Li using microorganisms72.
Microorganisms have mechanisms of resistance25,38,42,62 in order to survive in adverse environments, such as metal concentration above acceptable levels. The oldest survival strategy is bacterial biofilm45,35. This conformation provides a significant advantage of protection against environmental fluctuations as well as humidity, temperature and pH, concentrating nutrients, among others. Thus, regulation of life depends on the microorganism and the involvement of cellular signal complex networks, in which various proteins that respond to different environmental signs are involved58. Moreover, the active efflux of metal from the cytoplasm to the periplasmic space is another mechanism described for metal resistance in gram negative bacteria. It is carried out by ATPases located in the internal membrane of bacteria62. In addition, when submitted to stress, cells form chemically reactive molecules containing oxygen such as peroxides, superoxides, hydroxyl radical and singlet oxygen called reactive oxygen species (ROS)20. The tricarboxylic acid cycle (TCA) is a metabolic network that acts both as a scavenger and generator of ROS, whose modulation appears to be an important strategy in O2-dependent organisms to regulate the intracellular levels of ROS. TCA cycle is an important metabolic network used by organisms to survive in an oxidative environment43.
The high-altitude Andean lakes (HAAL) are unique, not only for their geographical characteristics and wide range of extreme environmental conditions, but also for their abundant biodiversity3. The microbial communities that have evolved within these high-altitude aquatic ecosystems tolerate a wide range of chemical and physical stress, such as strong fluctuations in daily temperature, hypersalinity, and variable pH. They have adapted to high levels of UV radiation, a low level of nutrient availability, and high concentrations of heavy metals and metalloids3,80. Indigenous extremophiles have apparently developed efficient survival strategies and should be able to produce biomolecules adapted to their unusual living conditions that might represent valuable sources of novel bioproducts (e.g. antioxidants, pigments, extremoenzymes) as it has been shown for other extremophiles63.
The genus name “Rhodococcus” is described as aerobic, gram positive, non-motile, mycolate-containing, nocardioform actinomycetes29. The ubiquitous Rhodococcus bacteria display a large set of enzymatic capabilities and can be found in a widespread of environments9. Rhodococci have been isolated from a large variety of sources including soils, rocks, boreholes, groundwater, marine sediments, animal dung, insect guts as well as from healthy and diseased animals and plants29,33. Rhodococcus spp. is able to survive in the presence of high doses of toxic compounds, as well as under desiccation conditions, carbon starvation, a wide range of temperatures (from 4°C to 45°C), UV irradiation and osmotic stress5,41. Strains have been isolated from deep sea and sea level sediments, alpine soils, the Arctic and the Antarctic19. Rhodococcus may play a significant role in the bioaccumulation of heavy metal ions. Two Rhodococcus strains were found to accumulate high levels of cesium ions (5% and 9% of the dry biomass)71. Moreover, Prasad et al. (2011) reported that the biomass derived from Rhodococcus sp. WB-12 cells have the potential to be used as biosorbent for the removal of arsenic from contaminated water56.
The Rhodococcus genus has a large set of enzymes participating in several biocatalytic and degradation reactions with industrial relevance9,19,39. The versatile metabolism conferred by their extended array of enzymes and the resistance displayed by Rhodococci makes these bacteria potential candidates to be used in environmental and biotechnological applications9,70,73.
Based on the above mentioned information, the aim of this study is to characterize the behavior of Rhodococcus sp. A5wh in order to understand the strategy followed by this strain in response to the presence of Li.
Materials and methodsBacterial strain and culture conditionsThe strain used in this research was obtained from the LIMLA-PROIMI Extremophilic Strain Collection. Rhodococcus sp. A5wh (white-mutant) is a spontaneous mutant detected after UVR-treatment of the original wild-type strain, Rhodococcus sp. A5Wt (red original) which was previously isolated from Laguna Azul10. Rhodococcus sp. A5wh was identified by the sequencing of 16S rRNA (accession number DQ112024.2). It was selected based on the results of a previous study that reported its capacity of growing in the presence of lithium and also its tolerance of brine conditions. The strain was routinely grown in Luria-Bertani broth (LB, Britania) at 30°C.
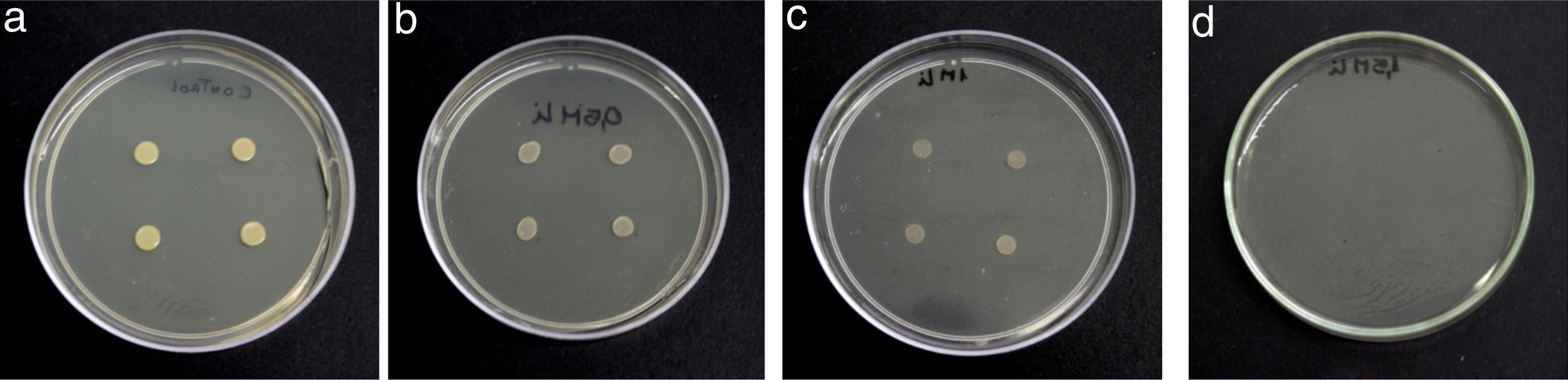
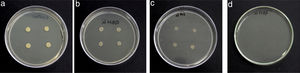
Semiquantitative screening to metal resistanceTo determine Li tolerance, the assays were carried out in a Petri dish containing LB agar supplemented with ions Li+ ions, added as LiCl (Sigma–Aldrich, Germany). Adequate amounts of metal salt were added to the culture medium to obtain a final concentration of Li equal to 0.7, 1 and 1.5M. LB agar+Li+ plates were inoculated with four drops of 10μl each of bacterial suspension (OD600nm=0.06) and incubated at 30°C for 48h for growth observation.
Lithium accumulation experimentsTo assay the ability to accumulate Li, the experiments were conducted as follows: resting microorganisms (0.28g dry weight) were suspended in a 20ml of solution containing 0.1M lithium supplied in the form of LiCl; the suspensions were shaken for 2h at room temperature at 70rpm. The pH of the solutions was adjusted to 6.0 using 0.1M HCl. The samples were centrifuged at 8000rpm (Beckmann) for 15min. The amounts of metal accumulated by the cells were determined by subtracting the Li ion concentration remaining in the supernatant from the original concentration of 0.1M. The metal concentration was measured using inductively coupled plasma optical emission spectrometry (ICP-OES) (Perkin Elmer).
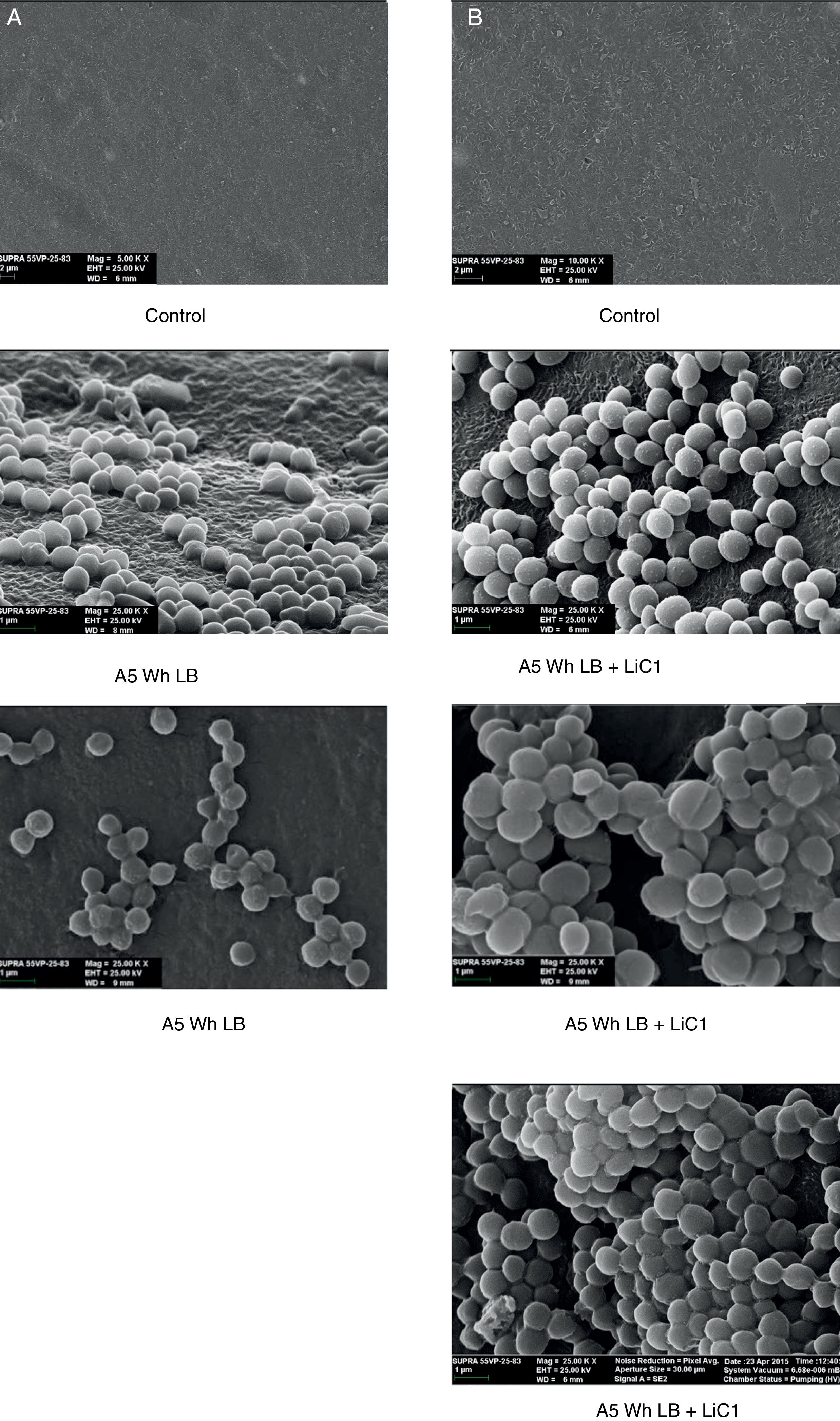
Biofilm formationFor the examination of biofilm development, cultures in the absence and presence of 1M Li were performed. Polystyrene pieces (5mm×2mm) were selected as a support matrix. They were placed in tubes containing 30ml of LB and LB medium with 1M of Li. Tubes were inoculated and incubated at 30°C with shaking at 170rpm during five days. After incubation, bacteria were separated by centrifugation (10000rpm) for 15min at 4°C, washed twice with distilled water. The samples were fixed and observed by scanning electron microscopy (SEM) using a SEMZeiss Supra 55vp at the Centro Integral de Microscopía Electrónica (CIME-CONICET-UNT).
Two-dimensional electrophoresis (2DE)For the proteomic assays a single colony of Rhodococcus sp. A5wh was grown in LB broth at 30°C for 24h and this culture was used for further inoculation of 500ml of LB (control), and 500ml of LB supplemented with 1M Li at initial OD600nm 0.1. Erlenmeyer flasks were incubated until the mid-exponential growth phase (OD600nm 1.5±0.05), and cells were harvested by centrifugation (7000rpm for 10min at 18–20°C) and washed four times with Tris–HCl buffer (0.1M) pH 7.5 and the pellets resuspended in 5ml of the same buffer. A French press at 2000psi (SLM Instruments, Inc., Haverhill, MA, USA) was used to break the cells. Unbroken cells and other debris were removed by centrifugation at 14000rpm for 10min at 4°C (Beckman, USA). Total protein concentration was determined by the Bradford method using BioRad reagents (Biorad, Richmond, CA), and bovine serum albumin (Sigma) as standard. Aliquots of 450μg were stored at −80°C until the isoelectrofocusing assay. Three independent assays for each condition were performed. Sample preparation and 2DE gels were carried out according to Belfiore et al.8 with some modifications. Samples containing 450μg of protein of Rhodococcus sp. A5wh (200μl) were treated with 2μl of bezonase (Novagen®) at 37°C for 30min and 3 volumes of cold acetone were added for protein precipitation. After incubation at −20°C for 16h, samples were centrifuged (14000rpm, 10min), the supernatant was carefully removed and the protein air-dried. Proteins were then solubilized in 40μl of solubilization mixture, centrifuged for 10min at 3500rpm, and loaded onto immobilized pH gradient strips (pH 4.0–7.0, 18cm, GE Health Care, Sweden). Gels were passively rehydrated for approximately 20h. The isoelectrofocusing assay was performed using IPGphor (GE, HealthCare, Sweden) at 53500V/h and the focusing strips were stored at −20°C until the second dimension was performed. The second dimension was performed by SDS-PAGE on gels containing 12.5% (w/v) polyacrylamide and carried out in a Bio-Rad Protean II xi cell (Biorad, Richmond, CA): proteins were resolved overnight at a constant current of 11mA/gel at 4°C. Gels were stained with Biosafe colloidal Coomassie blue (Biorad, Richmond, CA, USA) according to the manufacturer's instructions, scanned with Image Scanner III and analyzed with Prodigy SameSpots (Nonlinear Dynamic Group, UK). For each growth condition, at least three biological replicates were performed.
Protein identification using peptide mass fingerprintingDigitalized images (300dpi) of stained gels were aligned using the Prodigy SameSpote software version 1.0.3400.25570 (Nonlinear Dynamics Group, UK). Briefly, prominent spots were used to manually assign vectors in each gel image and the automatic vectors feature of the software was used to add additional vectors, which were manually verified. These vectors were used to wrap the images and align the spot position to a common reference gel. Spot detection performed according to this reference gel was edited and artifacts were removed. To correct the variability due to staining and to reflect the quantitative variation in intensity of protein spots, the spot volumes were normalized as a percentage of the total volume in all spots in the gel. A spot was considered significant when its resulting normalized volume showed more than 1.2-fold variation with respect to the control (LB) at the level of p<0.05. At least 3 biologically independent replicates for each growth condition were performed to obtain 3 gels for each assay.
Individual spots were excised from gels and the mass spectrometry analyses were carried out by CEQUIBIEM (Centro de Estudios Químicos y Biológicos de Espectrometría de Masa), Facultad de Ciencias Exactas y Naturales, UBA, Argentina. To determine protein identity, the MASCOT (Matrix Science Inc., Boston, MA; http://www.matrixsciende.com/search-form-select.html) program was used to identify proteins from peptide mass fingerprints. Fragmentation was carried out with the more intense MS peaks (MS/MS). When possible, MS and MS/MS information were combined for one or more peptide searches.
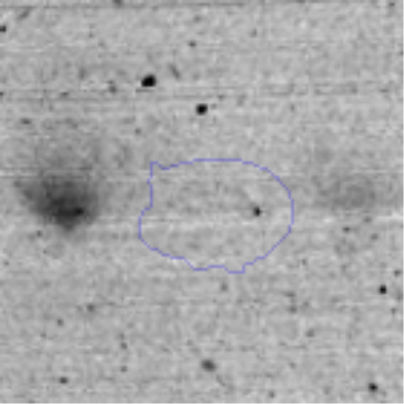
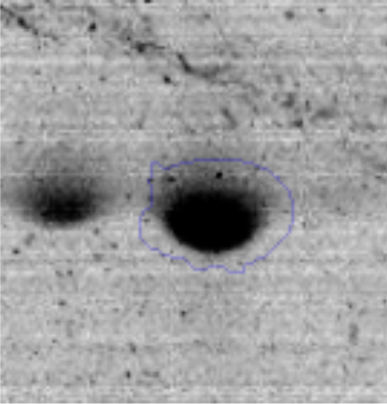
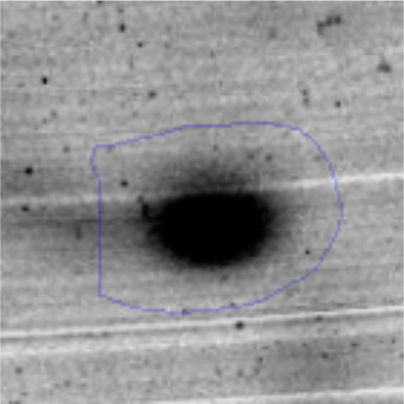
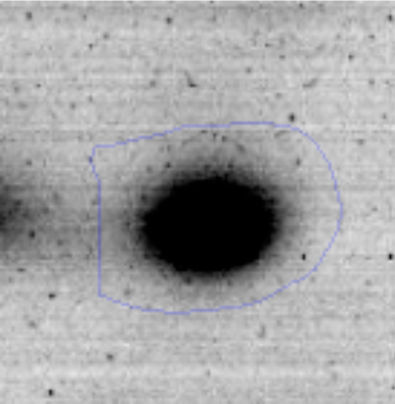
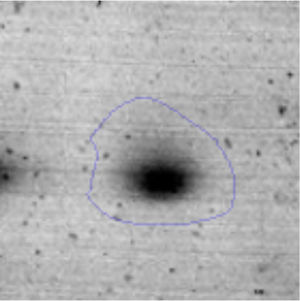
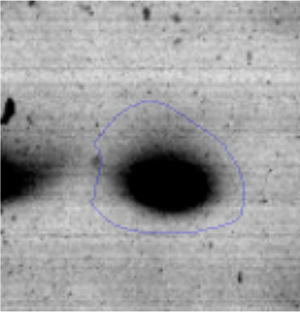
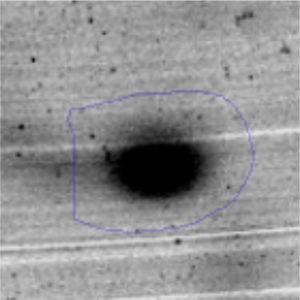
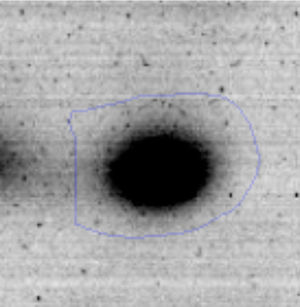
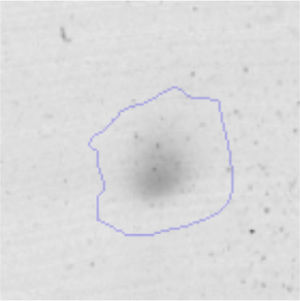
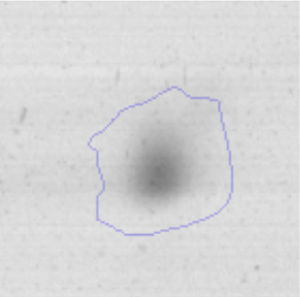
Results and discussionRhodococcus sp. A5wh was isolated from Laguna Azul, an oligotrophic lake, located at 4450m above sea level in Catamarca, Argentina, belonging to the extremophile culture collection of high-altitude Andean Lakes, isolated and described by Zenoff et al.80 With regard to cell viability, it was practically maintained unchanged despite the high concentration of Li (1M), however at 1.5M growth it was no longer detected on the plate (Fig. 1).
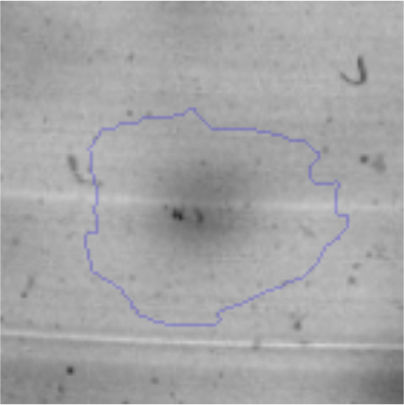
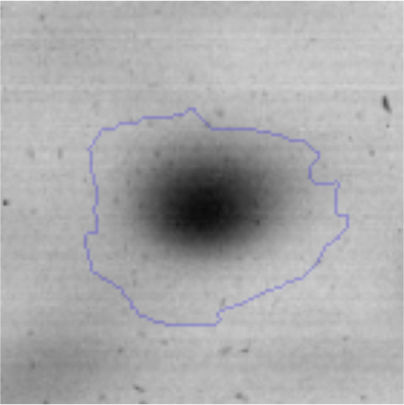
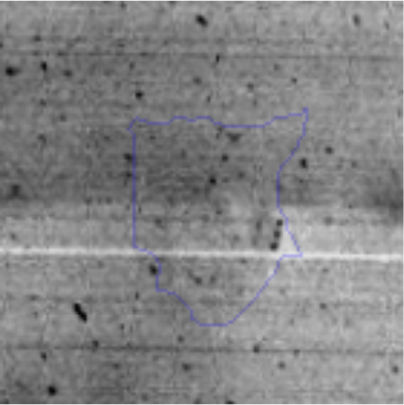
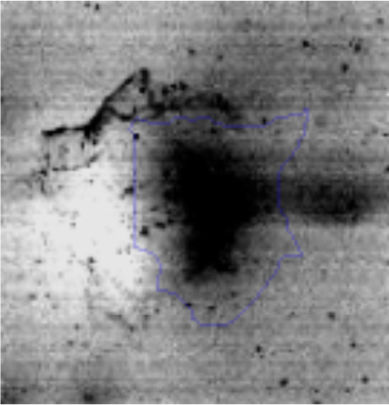
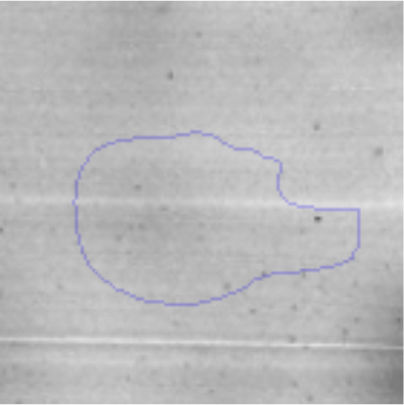
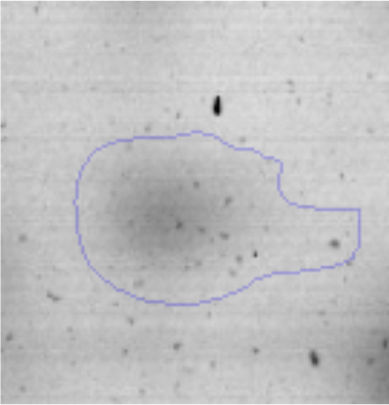
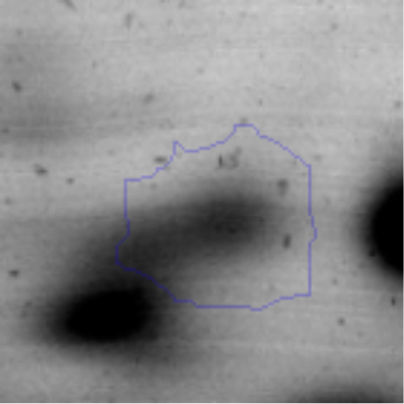
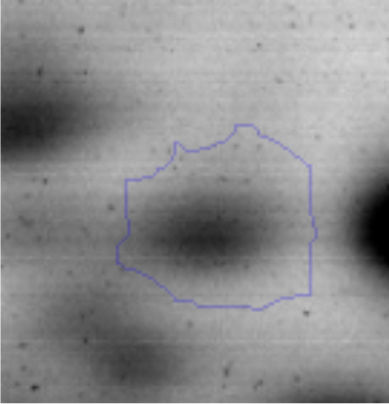
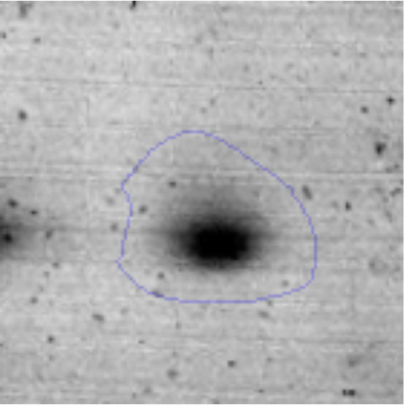
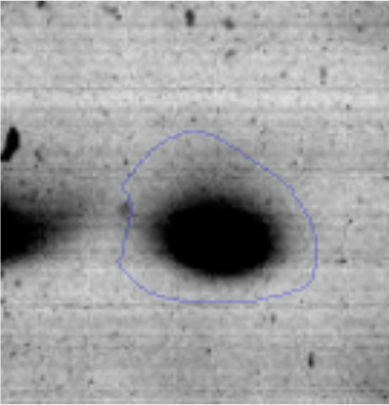
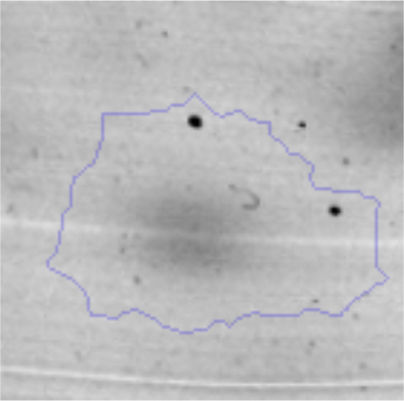
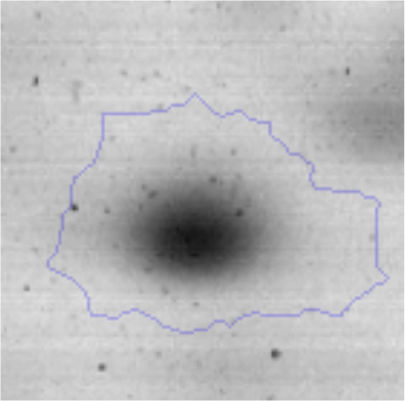
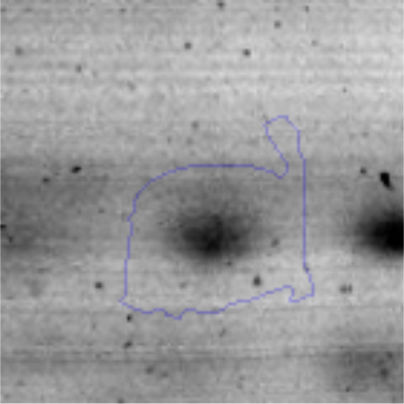
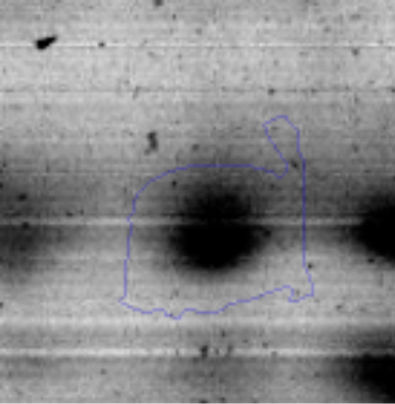
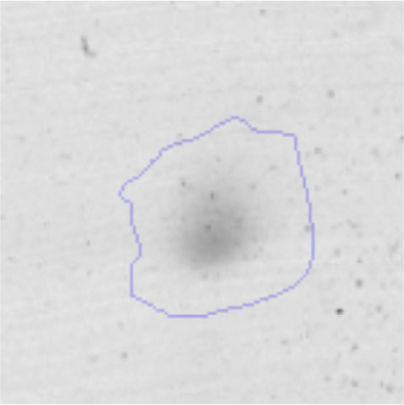
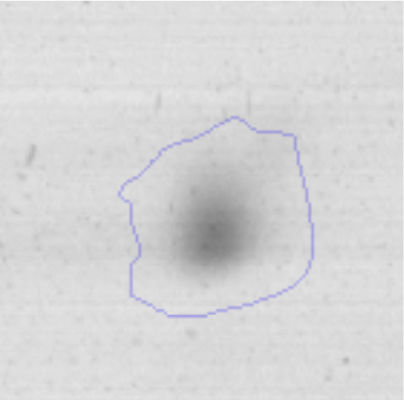
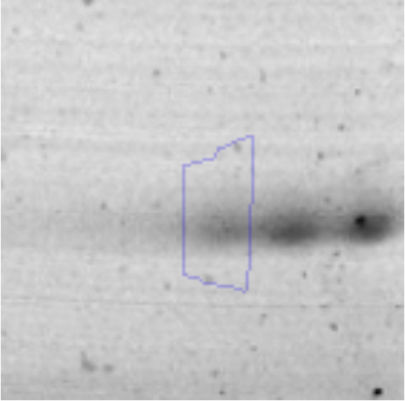
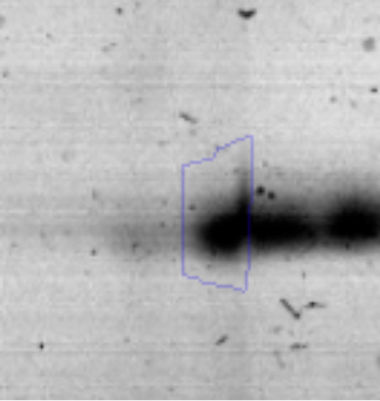
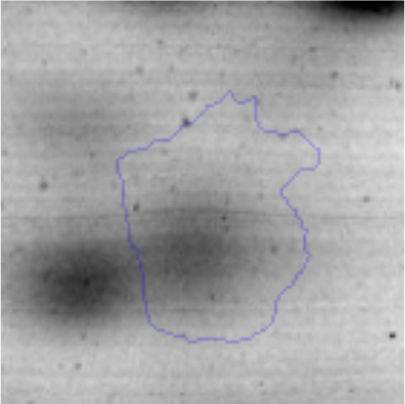
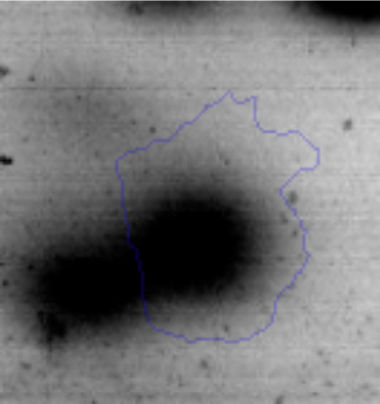
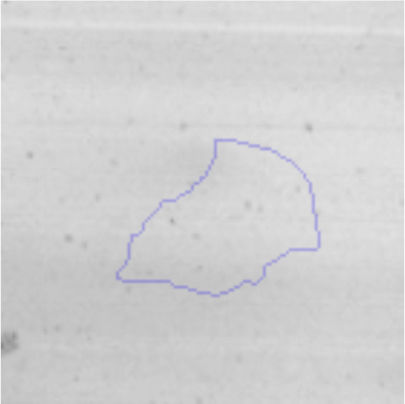
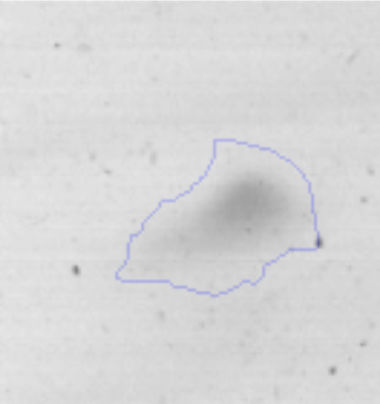
The microscopic examination of the biofilm revealed, as shown in Figure 2, that Rhodococcus sp. A5wh forms a biofilm in the presence and absence of Li, using polystyrene as support. Nevertheless, in the presence of this metal, bacterial aggregations are more dense and they cover more surface area than the other biofilms. This result would be expected since biofilms have been found to protect the microbial community from environmental stresses2,24.
In the accumulation assays performed, it was found that Rhodococcus sp. A5wh was able to accumulate 39.52% of Li/g microbial cells in 180min. Generally, the teichoic acid polymers of gram positive bacteria confer a strong negative charge on the surface of the cell wall because of their high content of ionized phosphate groups17. This result is in agreement with those reported by Tsuruta72, who determined that Li can be easily recovered using gram positive bacteria.
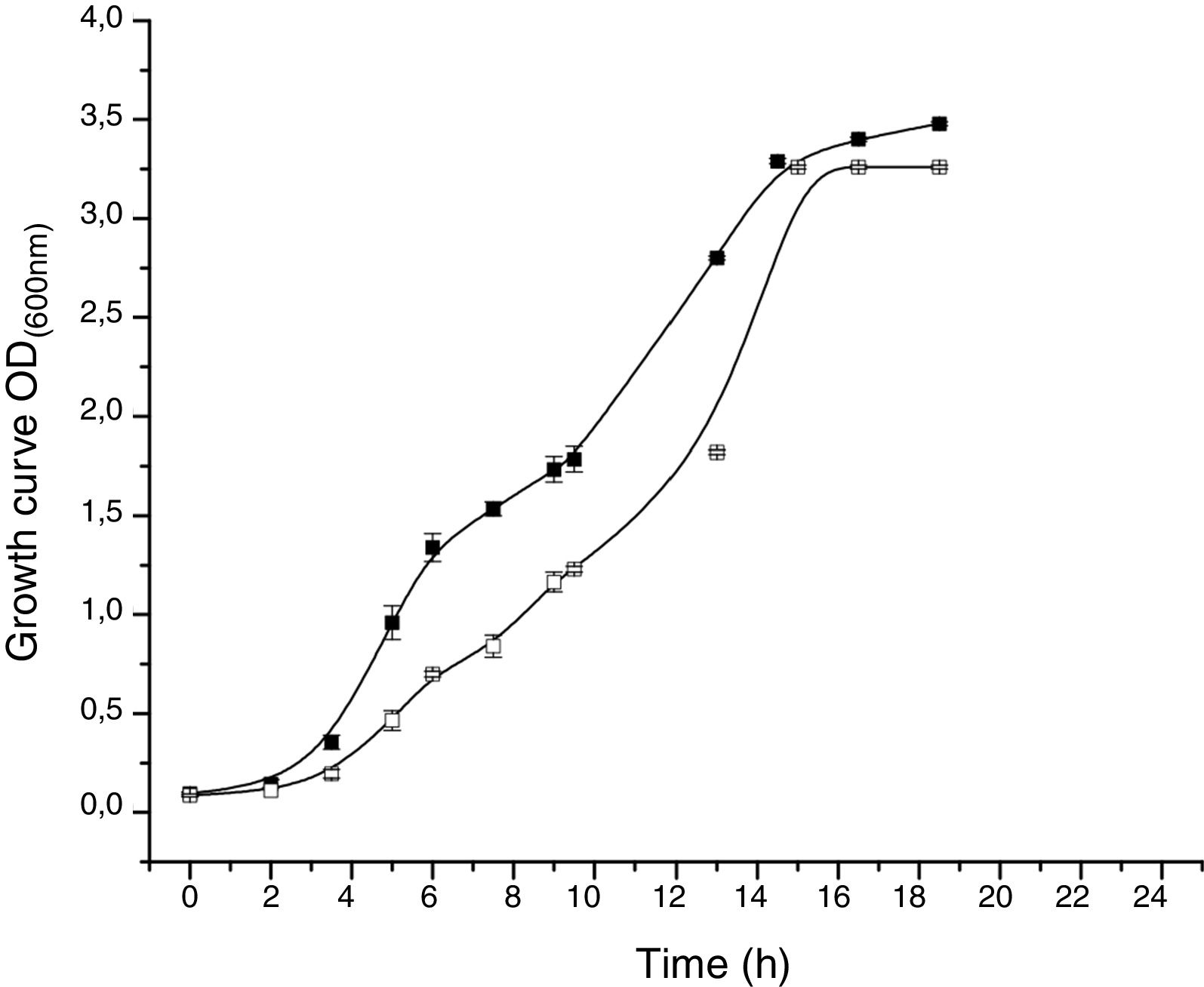
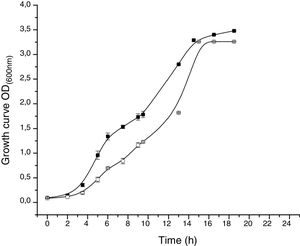
The proteomic response to Li was analyzed to identify the proteins that could be involved in this metal resistance in Rhodococcus sp. A5wh. Cells were grown in LB and LB+LiCl 1M (the highest concentration in which good growth is obtained) and harvested at log phase of growth (OD600mn 1.5). The presence of Li produced a slight retardation of growth. At 15h approximately, both cultures reached stationary phase (Fig. 3).
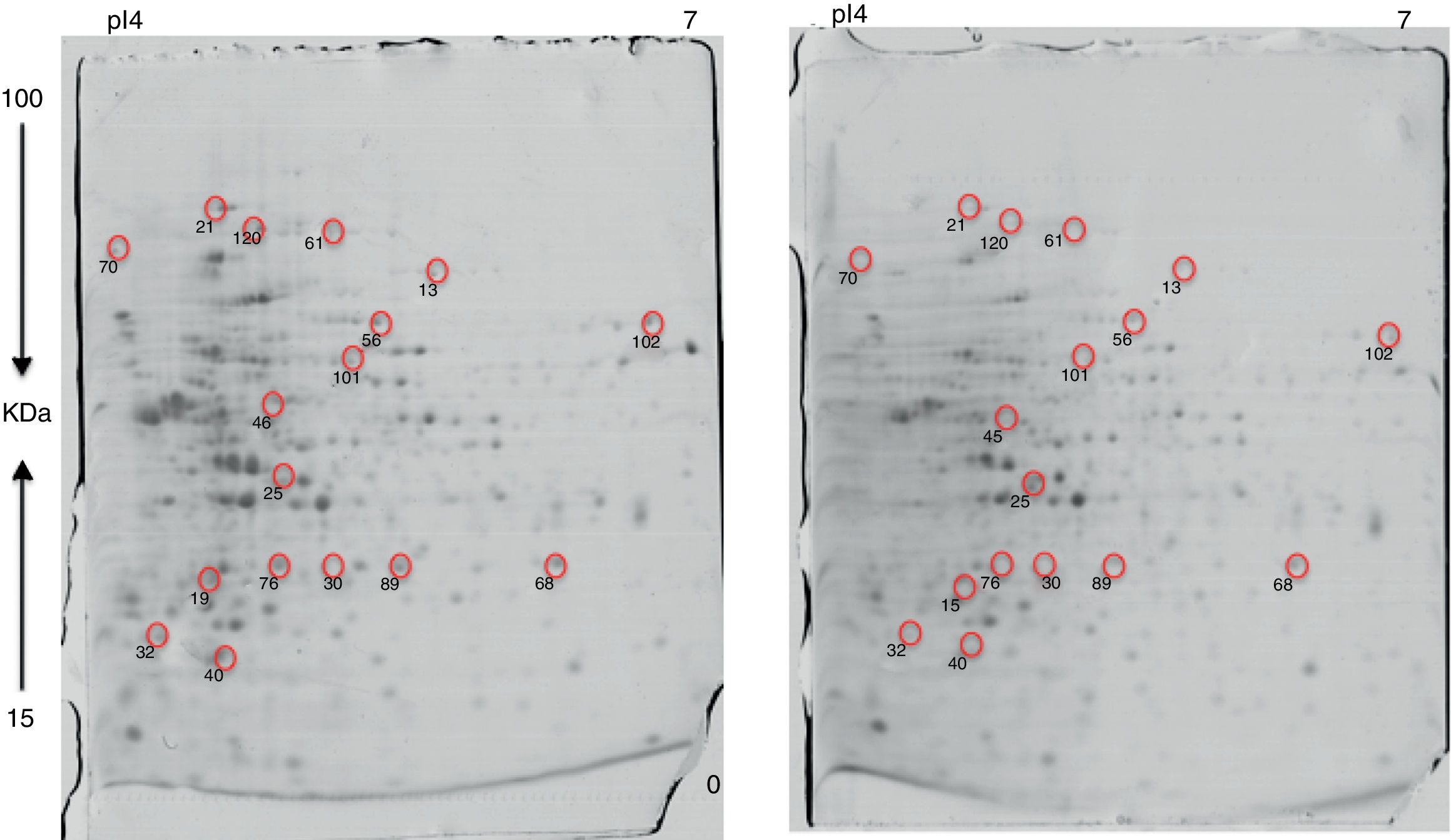
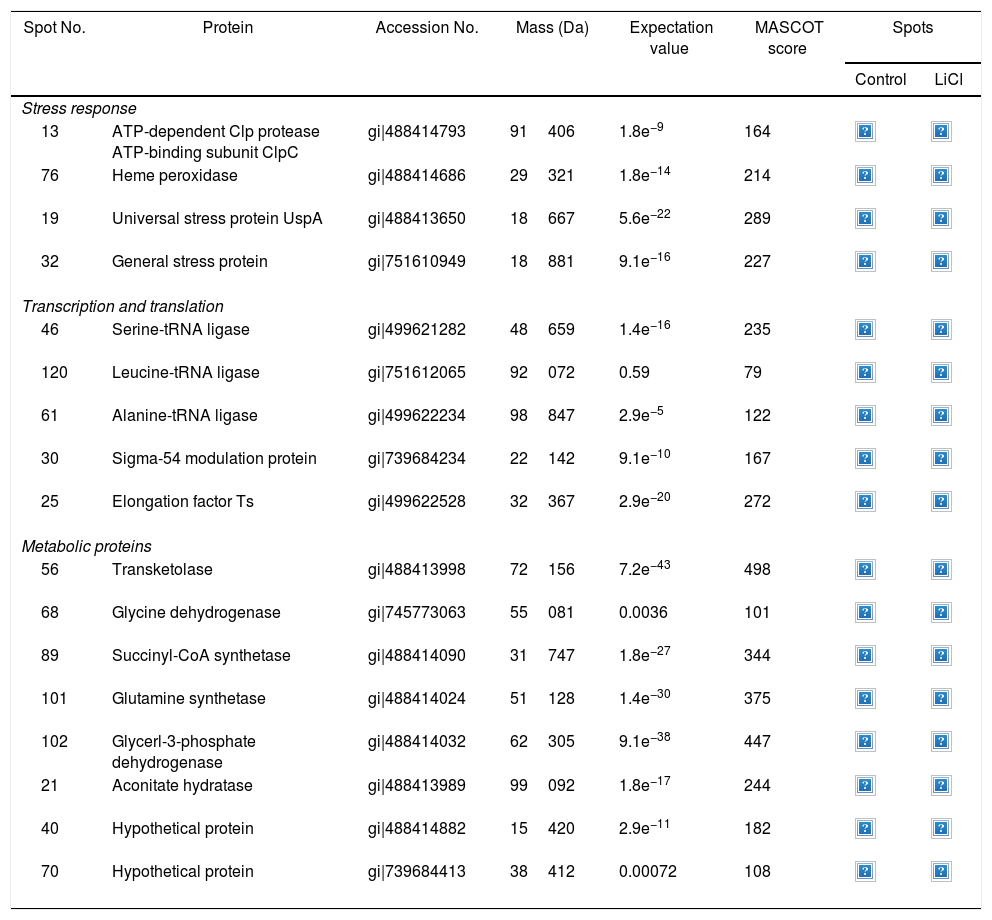
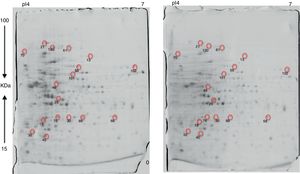
Approximately 146 spots expressed in a differential way under the assayed conditions were revealed in the gels (Fig. 4). Seventeen (17) statistically significant spots (Table 1) were identified by a factor greater than 1.2, using MALDI-TOF mass spectrometry.
Identification of differentially regulated proteins responding to lithium stress by Rhodococcus sp. A5wh
| Spot No. | Protein | Accession No. | Mass (Da) | Expectation value | MASCOT score | Spots | |
|---|---|---|---|---|---|---|---|
| Control | LiCl | ||||||
| Stress response | |||||||
| 13 | ATP-dependent Clp protease ATP-binding subunit ClpC | gi|488414793 | 91406 | 1.8e−9 | 164 | ||
| 76 | Heme peroxidase | gi|488414686 | 29321 | 1.8e−14 | 214 | ||
| 19 | Universal stress protein UspA | gi|488413650 | 18667 | 5.6e−22 | 289 | ||
| 32 | General stress protein | gi|751610949 | 18881 | 9.1e−16 | 227 | ||
| Transcription and translation | |||||||
| 46 | Serine-tRNA ligase | gi|499621282 | 48659 | 1.4e−16 | 235 | ||
| 120 | Leucine-tRNA ligase | gi|751612065 | 92072 | 0.59 | 79 | ||
| 61 | Alanine-tRNA ligase | gi|499622234 | 98847 | 2.9e−5 | 122 | ||
| 30 | Sigma-54 modulation protein | gi|739684234 | 22142 | 9.1e−10 | 167 | ||
| 25 | Elongation factor Ts | gi|499622528 | 32367 | 2.9e−20 | 272 | ||
| Metabolic proteins | |||||||
| 56 | Transketolase | gi|488413998 | 72156 | 7.2e−43 | 498 | ||
| 68 | Glycine dehydrogenase | gi|745773063 | 55081 | 0.0036 | 101 | ||
| 89 | Succinyl-CoA synthetase | gi|488414090 | 31747 | 1.8e−27 | 344 | ||
| 101 | Glutamine synthetase | gi|488414024 | 51128 | 1.4e−30 | 375 | ||
| 102 | Glycerl-3-phosphate dehydrogenase | gi|488414032 | 62305 | 9.1e−38 | 447 | ||
| 21 | Aconitate hydratase | gi|488413989 | 99092 | 1.8e−17 | 244 | ||
| 40 | Hypothetical protein | gi|488414882 | 15420 | 2.9e−11 | 182 | ||
| 70 | Hypothetical protein | gi|739684413 | 38412 | 0.00072 | 108 | ||
In the presence of lithium, 12 proteins (19, 21, 25, 32, 40, 46, 56, 68, 76, 89, 101, 102) were overexpressed, while five (13, 30, 61, 70, 120) were found only in the presence of this metal (absent in the control). At the moment, there is no information on the pathways for Li resistance49, for this reason we consider that our results are important to understand the mechanisms followed by Rhodococcus sp. A5wh.
Rhodococcus sp. A5wh showed overexpression of UspA (spot 19) in the presence of Li. This protein is important for the survival during cellular growth arrest, but the exact physiological role of the Usp proteins is not known. Nachin et al.48 described that UspA and UspD are involved in regulating cell capacity to withstand oxidative agents in Escherichia coli. In a previous work, Nyström and Neidhart50 also showed that UspA is important for H2O2 resistance during growth in E. coli. The induction of this protein is associated with the stress response. Patrauchan et al.55 reported the expression of Usp in Rhodococcus jostii RHA1 under carbon starvation. However, there is no information about the adaptative response of Rhodococcus against Li.
Spot 76 identified as heme peroxidase, resulted overexpressed in the presence of lithium. This protein catalyzes a number of oxidative reactions. Moreover, under the presence of Li, the increase of the ATP-dependent Clp protease ATP-binding protein (spot 13) is not surprising, since the cell is in a stressful situation. ClpP proteases degrade short-lived regulatory proteins as well as misfolded and damaged proteins that occur more often under stress23, thus maintaining cellular homeostasis79. Finally, the general stress protein (spot 32) was detected in the presence of Li; it is probably necessary for maintenance in order for cell survival.
In addition, the differential proteome of Rhodococcus sp. A5wh revealed the overexpression of proteins related to transcription and translations processes, as for example elongation factor protein Ts (spot 25), serine tRNA ligase (spot 46) and leucine tRNA ligase (spot 120). Sigma-54 modulation protein (spot 30) and alanine tRNA ligase (spot 61) were expressed only in the lithium condition.
EF-Ts is the protein responsible for mediating the regeneration of EF-Tu–GDP complex, catalyzing the addition of aminoacyl-tRNA into the ribosome, enabling protein synthesis; in E. coli it has been proposed to have an additional role as a chaperone for denatured and unfolded proteins15. According to Caldas et al.16, the EF-Tu has a dual function in protein synthesis and stress response. The amount of ribosomal sub-units increases possibly due to the requirement of increased protein biosynthesis in order to cope with cellular stress reflecting the ability of Rhodococcus sp. A5wh to adapt and survive in the presence of lithium.
Additionally, another group of proteins overexpressed in the presence of Li belong to metabolic proteins (spots 21, 56, 68, 89, 101 and 102).
Spot 102 was identified as glycerol-3-phosphate dehydrogenase (GPDH) that is an enzyme that catalyzes the reversible redox conversion of dihydroxyacetone phosphate to glycerol 3-phosphate.
GPDH serves as a major link between carbohydrate metabolism and lipid metabolism76, and it is also a major contributor of electrons to the electron transport chain. This result suggests that the cell requires energy under stress. Albertyn et al.4 report that in Saccharomyces cerevisiae, GPDH is essential for growth under osmotic stress through the production and intracellular accumulation of glycerol as compatible solute. Here it may play the same role in the presence of LiCl.
It is known that TCA is a crucial component of respiratory metabolism26 and a major metabolic hub of the cell; the TCA also provides intermediates for biosyntheses11. Rhodococcus has a functional TCA in both directions (oxidative and reductive) according to the conditions6. It is probable that this strategy allows the cells to respond rapidly to environmental changes as our results revealed overexpression of certain enzymes involved in this pathway such as aconitate hydratase (spot 21) and succinyl CoA synthetase (spot 89). The first enzyme catalyzes the isomerization of citrate to isocitrate, while the succinyl-CoA synthetase catalyzes the formation of succinate and CoA from succinyl-CoA with simultaneous ATP formation. This cycle is a source of biosynthetic precursors, its metabolites are important for biosynthetic intermediate of many other cellular components: lipids, carbohydrates, amino acid and nitrogenous bases. Hamel and Appanna30 described that oxalic acid plays a pivotal role in the adaptation of Pseudomonas fluorescens to aluminum stress. They report that the TCA cycle is tailored in order to generate the necessary precursor for oxalate synthesis as a consequence of stress. On the other hand, Singh et al.68 observed that P. fluorescens increased the activity of succinyl-CoA synthetase in the stress aforementioned route to ATP synthesis. According to our results, it is probable that Rhodococcus carries out the same strategy to resist the stress caused by Li. The metabolism of Rhodococcus has evolved to adapt to a wide range of nutritional conditions. This adaptation often involves the flexibility of the central metabolism, which usually provides energy and precursors for the biosynthetic processes, either during growth or during non-replicative metabolically active periods.
Otherwise malate: quinone oxidoreductase (MQO), the last step of the TCA cycle consists of the regeneration of oxalacetate by the oxidation of malate. MQO is responsible for the flux from malate to oxalacetate in the TCA cycle. Molenaar et al.47 showed that MQO-deficient mutants were unable to grow. These results indicate that this enzyme is essential for a functional TCA cycle in Corynebacterium glutamicum.
The glyoxylate cycle acts as an anaplerotic route that allows normal operation of the TCA cycle. Spot 68 was identified as glycine dehydrogenase, an enzyme that catalyzes the reductive amination of glyoxylate to glycine; this reaction may represent a significant alternate pathway for the formation of glycine in the absence of transamination.
Microorganisms have developed several strategies of detoxification. One of the most powerful adaptive strategies involves intracellular accumulation of small organic compounds, the so-called compatible solutes, which equilibrate cellular osmotic pressure14,78, this being regarded as a protein synthesis-independent mechanism. These osmolytes such as glycine, proline, glutamate, glutamine, trehalose, and glycerol may also protect native proteins from denaturation and favor the formation of native protein oligomers, some of these behaving as “chemical chaperones” by promoting the correct refolding of unfolded proteins in vitro and in the cell21. Here we have demonstrated that the overexpression of glutamine synthetase and glycine dehydrogenase are a salt-induced process in Rhodococcus; it is evident that it could act as a mechanism of adaptation (osmoprotector) to the environment with LiCl.
In addition, glutamine synthetase (spot 101) was overexpressed in the presence of Li. This enzyme is in charge of catalyzing the condensation of glutamate and ammonia to form glutamine69. Glutamine plays a role in a variety of functions such as protein and purine synthesis, and like glutamate, it acts as carbon source to recharge the citric acid cycle. It is reported that glutamine, which can be synthesized from glutamate, increases in concentration in cultures of E. coli46 and Salmonella typhimurium18, under osmotic stress conditions. Saum et al.64 observed that glutamine synthetase activity was readily detectable in Halobacillus halophilus and its induction was strictly salt-dependent, reaching a maximum of 300% stimulation at 3.0M NaCl. Furthermore, glutamine has been reported as a compatible solute in the members of the genus Corynebacterium27 which is closely related to Rhodococcus and, as a result, could be working as osmoprotector in the presence of Li.
It is important to mention that the proteins involved in biofilm formation were not identified in this work. This may happen because Rhodococcus sp. A5wh is able to produce them in both conditions (with or without Li); and for this reason, they were not found as differential spots (Fig. 2). Moreover, the pump efflux (Na+/K+) is another defense mechanism that has been described for prokaryotic and eukaryotic cells and has not been detected in the proteomic differential screen. Lithium belongs to the group of alkali metals together with sodium, potassium, rubidium and cesium. It is probable that sodium is replaced by lithium due to similar structures. Membrane proteins are often highly insoluble and therefore, not readily resolved by 2D-PAGE7. This may be the reason why we have no results about these proteins. However, two hypothetical proteins with unknown function were identified.
Finally, transketolase (spot 56) is a key enzyme in the non-oxidative branch of the pentose phosphate pathway that transfers a two-carbon glycoaldehyde unit from ketose-donor to aldose-acceptor sugars. It participates in the formation of xylulose 5-phosphate for NADPH production. This up-regulation could indicate that more NADPH was generated under Li conditions. This coenzyme is involved in the synthesis of nucleic acids and lipids. Unlike our results, Pandey et al.54 and Belfiore et al.8 observed that the synthesis of this protein was decreased under arsenic stress. This could be due to high sensitivity to H2O237 accumulated in the cells after the treatment with As and/or competitive inhibition of the catalyzed reactions by the metalloid. These results constitute another evidence of the adaptive response of Rhodococcus against Li stress, in order to guarantee their survival.
ConclusionThis report provides novel insights into the pathway involved in the Li response of Rhodococcus sp. A5wh. In conclusion, seventeen proteins were successfully identified. The overexpression of stress proteins, proteins related to the TCA, and enzymes involved in the synthesis of compounds that can be used as osmoprotectants are an important contribution to understanding the mechanism used by Rhodococcus sp. A5wh to guarantee adaptation to the environment.
Although Rhodococcus sp. A5wh was able to absorb Li, more assays are needed in order to exploit the potential of strains in terms of their biotechnological applications such as their use in biomining.
Ethical disclosuresProtection of human and animal subjectsThe authors declare that no experiments were performed on humans or animals for this study.
Confidentiality of dataThe authors declare that no patient data appear in this article.
Right to privacy and informed consentThe authors declare that no patient data appear in this article.
Conflict of interestThe authors declare that they have no conflicts of interest.
This work was supported by Préstamo BID PICT 2010 N°1788 – Agencia Nacional de Promoción Científica y Tecnológica. We thank Dr. S. Fadda for her generosity.