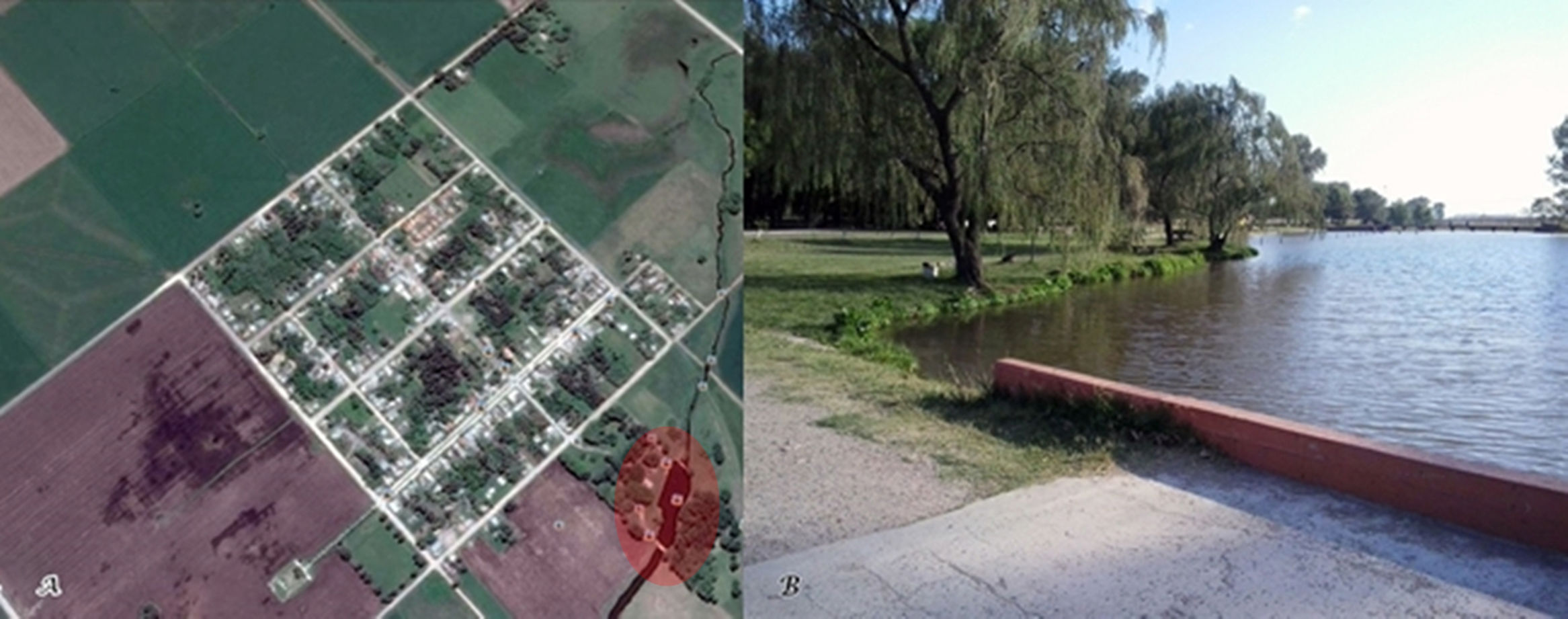
Ten Leptospira spp. strains were isolated from water samples from Nievas stream, Olavarría, Buenos Aires province (Argentina). The isolates showed the typical motility and morphology of the genus Leptospira under dark field microscopy, developing in liquid EMJH medium after eight days of incubation at 13°C and 30°C. All isolates were negative by the Multiple Locus Variable Number Tandem Repeat Analysis (MLVA). Molecular identification by 16S rRNA gene sequencing identified all isolates as nonpathogenic leptospires. Four isolates showed a genetic profile identical to that of the reference strain Leptospira biflexa serovar Patoc, and six isolates revealed sequence similarities within the 97–98% range, closely related to Leptospira yanagawae and Leptospira meyeri, respectively. Strains ScialfaASA42, ScialfaASA45, ScialfaASA44, ScialfaASA47, ScialfaASA49, ScialfaASA50 and ScialfaASA51 possibly represent a novel species of the genus Leptospira.
Se aislaron 10 cepas de Leptospira spp. a partir de muestras de agua del arroyo Nievas, partido de Olavarría (provincia de Buenos Aires, Argentina). Los aislamientos mostraron motilidad y morfología típica del género Leptospira bajo microscopía de campo oscuro y se desarrollaron en medio líquido EMJH después de 8 días de incubación a 13 y 30°C. Todos los aislamientos fueron negativos por MLVA, y mediante la secuenciación del gen 16S del ARNr se identificaron como leptospiras no patógenas. Cuatro de estos aislamientos mostraron un perfil genético idéntico a la cepa Leptospira biflexa serovar Patoc de referencia, en tanto que 6 de ellos presentaron similitudes de secuencias estrechamente relacionadas con las especies Leptospira yanagawae y Leptospira meyeri dentro del intervalo del 97 y 98%, respectivamente. Las cepas ScialfaASA42, ScialfaASA45, ScialfaASA44, ScialfaASA47, ScialfaASA49, ScialfaASA50 y ScialfaASA51 posiblemente representen una nueva especie del género Leptospira.
Leptospirosis is a bacterial zoonotic disease caused by pathogenic species of the genus Leptospira, which is mainly transmitted to humans by direct contact with rodents and domestic animals, mainly dogs and cattle. However, human outbreaks are frequently produced by contact with water that is contaminated with urine of animal reservoirs9,13; and leptospires penetrate abraded skin or mucous membranes and disseminate through the organism. The genus Leptospira includes at least 22 species arranged into three large subgroups based on 16S rRNA phylogeny, ten (10) pathogenic species, seven (7) saprophytic species and five (5) intermediate species2,6. In Argentina, there are few studies of leptospires in environmental sources, mainly from water.
In this paper, we report the circulation of two non-pathogenic species of the genus Leptospira isolated from water samples from Nievas stream, Olavarría, Argentina.
Initially, leptospiral strains were isolated from water samples from Nievas stream, Olavarría, Buenos Aires province, Argentina (Fig. 1) by following isolation procedures. Water samples were collected in a recreational area during March 2015, and transported in sterile 500ml glass bottles. Temperature and pH were monitored in the field. Water samples were filtered through a sterile membrane. In this study, a prefiltering technique was applied using Whatman filter paper before filtration through a 0.22μm pore size membrane filter. A sample of the filtered water (1ml) was inoculated into Ellinghausen–McCullough– Johnson–Harris (EMJH medium: Difco Laboratories, Detroit, Michigan, USA) liquid medium with the addition of 5-fluorouracil (200μg/ml) as selective antimicrobial agent. Cultures were incubated by duplicate at 13°C and 30°C for 90 days, and leptospiral growth was monitored weekly using dark field microscopy. On day eight, growth was observed at both temperatures, and ten strains were isolated.
DNA templates were obtained using Chelex Resin-100 (Bio Rad). Multiple Locus Variable Number Tandem Repeat Analysis (MLVA) was performed using two sets of oligonucleotides specific for pathogenic leptospires (Leptospira interrogans, Leptospira kirschneri and Leptospira borgpetersenii). Oligonucleotides that hybridized to the flanking regions of the VNTR4, VNTR7, VNTR9, VNTR10, VNTR19, VNTR23 and VNTR31 loci were used to discriminate strains of L. interrogans and oligonucleotides that hybridized to the flanking regions of the VNTR4, VNTR7, VNTR10; Lb4 and Lb5 loci were used for L. kirschneri, L. borgpetersenii and L. interrogans strains7,8. The final volume (50μl) of each reaction mixture contained polymerase chain reaction (PCR) buffer (20mM Tris–HCl, pH 8.4, 50mM KCl), 200μM deoxynucleoside triphosphates, 2μM each of the corresponding forward and reverse primers, 2mM MgCl2, 1.25U of Taq DNA polymerase (Invitrogen) and 5μl of DNA template. PCR amplifications were carried out in a Thermo Scientific PxE 0.2 Thermal Cycler, using the following cycling parameters: 94°C for 5min, followed by 35 cycles of denaturation at 94°C for 30s, annealing at 55°C for 30s and extension at 72°C for 90s, with a final cycle at 72°C for 10min. The amplified samples were examined by electrophoresis in ethidium bromide-containing 2% agarose gels in TAE buffer (40mM Tris-acetate, 1mM EDTA, pH 8.0) at 100V for 50min. Amplified DNA bands were visualized upon ultraviolet light exposure (Uvi Tec transiluminator BTS-20.M, Manufacturer UviTec, St. John's Innovation Centre, Cowley Road, Cambridge, England). Amplicon sizes were estimated using CienMarker (Biodynamics) and the GelAnalyzer 2010 program. To calculate repeat copy numbers, the following formula was used: number of repeats (bp)=[fragment size (bp)−flanking regions (bp)]/repeat size (bp). Repeat copy numbers were rounded down to the closest whole number. If the copy number was less than one, it was rounded to zero.
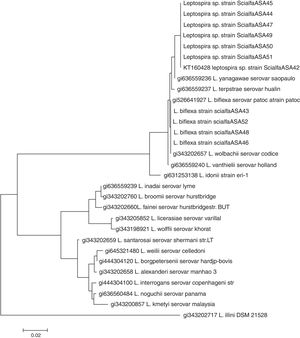
PCR targeting the 16S rRNA gene was carried out for bacterial identification. The following primers were used: 5′-GGCGGCGCGTCTTAAACATG-3′ and 5′-GTCCGCCTACGCACCCTTTACG-3′; these primers have the ability to amplify all pathogenic and nonpathogenic leptospires4. After verification of the amplicon by electrophoresis (in an ethidium bromide-containing 2% agarose gel) and visualization upon UV light exposure, PCR products were purified using a commercial kit (Embiotech). The sample was sequenced at the Institute of Biotechnology, National Institute of Agricultural Technology (Argentina) using a 3130xl Genetic Analyzer (Applied Biosystems). For alignment and construction of the phylogeny, the program MEGA version 5 was used12. The dendogram based on partial sequences of the 16S rRNA gene was constructed using Neighbor-joining with a bootstrap of 100 (Fig. 2).
Neighbor-joining analysis of the sequence 16S rRNA of all strains isolated. Phylogenetic analysis based of 16S rRNA including 19 representative species of Leptospira. The corresponding sequence of Leptonema illini strain DSM 21528 was used as an outgroup. The dendrogram was constructed using the Neighbor-joining method. Bootstrap values are displayed as percentages.
At the moment of water sample collection in Nievas stream, we observed a temperature of 24°C and pH 7.4. Growth was observed after 8 days in culture medium at 13°C and 30°C, and the isolates showed the typical motility and morphology of the genus Leptospira sp. (cells of 10–13mm in length and less than 0.22mm in diameter, characterized by flexuous motility) under dark field microscopy. Strains were maintained in Ellinghausen–McCullough–Johnson–Harris (EMJH) liquid medium.
All isolates were negative by the MLVA; however, molecular identification by 16S rRNA gene sequencing identified all isolates as non-pathogenic leptospires. Four isolates showed a genetic profile identical to that of the reference L. biflexa serovar Patoc (strains ScialfaASA43, ScialfaASA46, ScialfaASA48 and ScialfaASA52), and six isolates (strains ScialfaASA42, ScialfaASA45, ScialfaASA44, ScialfaASA47, ScialfaASA49, ScialfaASA50 and ScialfaASA51) with sequence similarities within the 97–98% range using Blast (results not shown), and closely related to species Leptospira yanagawae and Leptospira meyeri respectively. Strains ScialfaASA42, ScialfaASA45, ScialfaASA44, ScialfaASA47, ScialfaASA49, ScialfaASA50 and ScialfaASA51 possibly represent a novel species of the genus Leptospira. The sequence obtained from the strain ScialfaASA42 is available in GenBank under Accession number KT160428.
In Argentina isolates from water sources (rivers, lakes, and streams) have been obtained in Buenos Aires, Santa Fe, Corrientes and Jujuy province. In the period 1961–1996 a total of 47 saprophytes strains and 5 pathogenic strains were isolated from water (two unclassified L. interrogans, one L. interrogans serovar Icterohaemorrhagiae and one L. interrogans serovar Pomona)1. Brihuega et al.3 isolated a pathogenic strain from water samples from Reconquista river, which was classified as L. interrogans serovar Icterohaemorrhagiae strain Reconquista. In the year 2011, five strains were isolated from water samples from rural channels in Casilda, Santa Fe province, Argentina, and one was classified as L. borgpetersenii5. To the best of our knowledge, eight pathogenic Leptospira sp. strains have been isolated so far from environmental water sources in Argentina. In the center of Buenos Aires province, there have been no reports of pathogenic leptospires from water sources. However, in previous studies on wild animals captured in the region (Rattus norvegicus, Akodon azarae, Lycalopex griseus, Conepatus chinga and Didelphis albiventris), we showed the circulation of several strains belonging to L. interrogans serogroup Icterohaemorrhagiae: serovar Copenhageni strains Fiocruz L1-130 and M20, belonging to serovar Icterohaemorrhagiae RGA (in peri-urban and rural area). Furthermore, strains belonging to L. interrogans serogroup Canicola: serovar CanicolaHound Utrecht IV (in rural area) were found9–11. In the present study, we showed that in the selected environment, especially in water, conditions (pH and temperature) are optimal for the survival of non-pathogenic and pathogenic leptospires. In water samples from Nievas stream in Olavarria, Buenos Aires province, we observed the circulation of two saprophytics species of the genus Leptospira (L. biflexa serovar Patoc and Leptospira sp. with 97–98% sequence similarities to L. yanagawae and L. meyeri respectively). The MLVA assay and the molecular characterization of the 16S rRNA gene sequencing were important tools for the identification of these isolates as non-pathogenic leptospires; however, they were insufficient to define strains ScialfaASA42, ScialfaASA45, ScialfaASA44, ScialfaASA47, ScialfaASA49, ScialfaASA50 and ScialfaASA51 as known saprophytic species of the genus Leptospira. Further studies such as pulsed-field gel electrophoresis: PFGE; DNA–DNA hybridization, average nucleotide identity: ANI and genome-to-genome distance calculator, GGD, should be conducted to further study these isolated strains.
Ethical disclosuresProtection of human and animal subjectsThe authors declare that no experiments were performed on humans or animals for this study.
Confidentiality of dataThe authors declare that no patient data appear in this article.
Right to privacy and informed consentThe authors declare that no patient data appear in this article.
Conflict of interestThe authors declare that they have no conflicts of interest.