Clostridioides difficile is a spore-forming anaerobe microorganism associated to nosocomial diarrhea. Its virulence is mainly associated with TcdA and TcdB toxins, encoded by their respective tcdA and tcdB genes. These genes are part of the pathogenicity locus (PaLoc). Our aim was to characterize relevant C. difficile toxinotypes circulating in the hospital setting. The tcdA and tcdB genes were amplified and digested with different restriction enzymes: EcoRI for tcdA; HincII and AccI for tcdB. In addition, the presence of the cdtB (binary toxin) gene, TcdA and TcdB toxins by dot blot and the cytotoxic effect of culture supernatants on Vero cells, were evaluated. Altogether, these studies revealed three different circulating toxinotypes according to Rupnik's classification: 0, I and VIII, being the latter the most prevalent one. Even though more studies are certainly necessary (e.g. sequencing analysis), it is worth noting that the occurrence of toxinotype I could be related to the introduction of bacteria from different geographical origins.
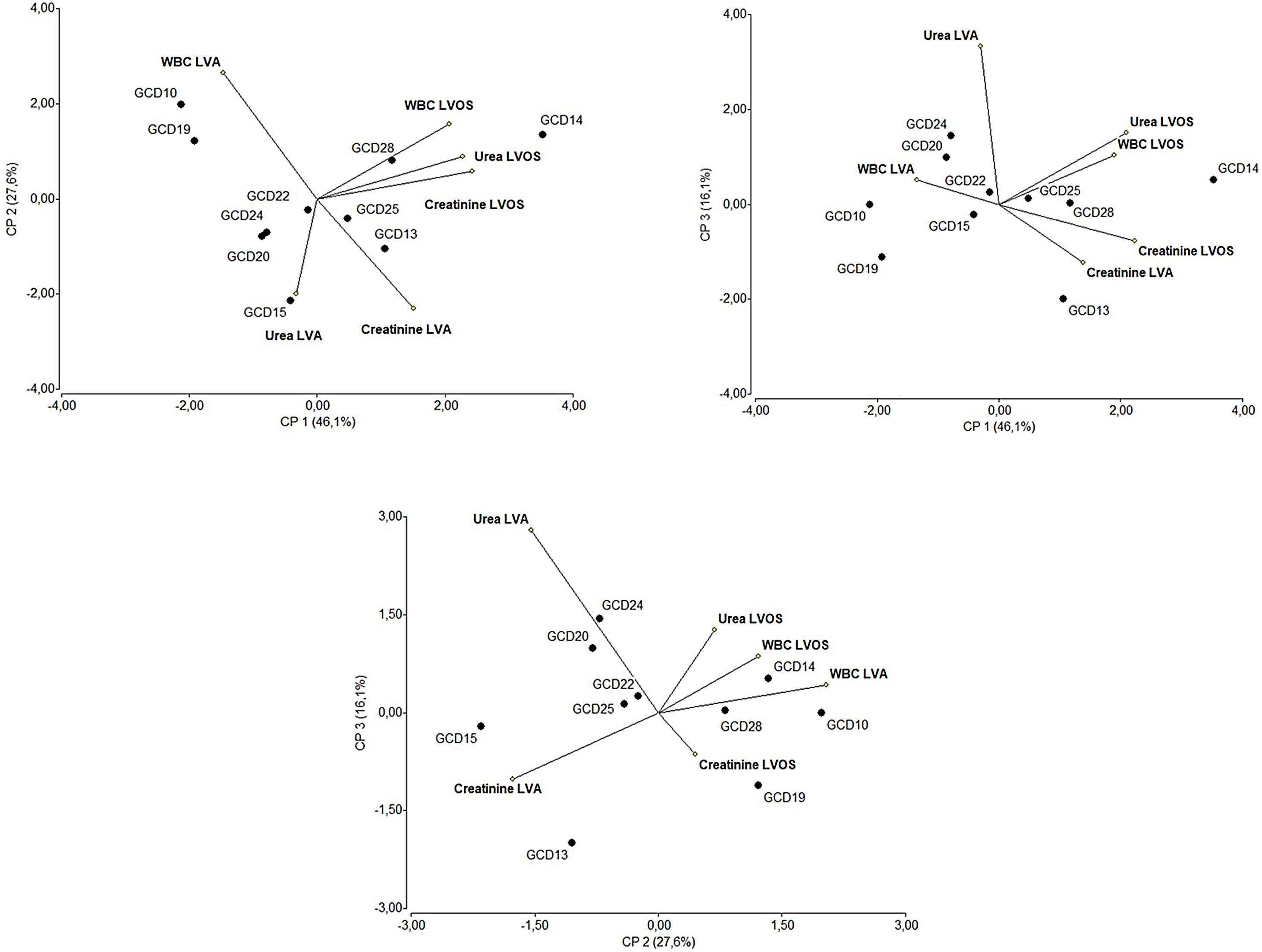
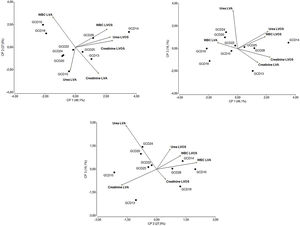
The multivariate analysis conducted on the laboratory values of individuals infected with the most prevalent toxinotype (VIII) showed that the isolates associated with fatal outcomes (GCD13, GCD14 and GCD22) are located in regions of the biplots related to altered laboratory values at admission.
In other patients, although laboratory values at admission were not correlated, levels of urea, creatinine and white blood cells were positively correlated after the infection was diagnosed.
Our study reveals the circulation of different toxinotypes of C. difficile strains in this public hospital. The variety of toxinotypes can arise from pre-existing microorganisms as well as through the introduction of bacteria from other geographical regions. The existence of microorganisms with different pathogenic potential is relevant for the control, follow-up, and treatment of the infections.
Clostridioides difficile es un anaerobio esporulado que se asocia con episodios de diarreas hospitalarias. Su virulencia se encuentra vinculada, principalmente, a las toxinas TcdA y TcdB, codificadas por sus respectivos genes, tcdA y tcdB, que son parte de un locus de patogenicidad (PaLoc). Nuestro objetivo fue caracterizar los toxinotipos de C. difficile circulantes en un hospital público. Los genes tcdA y tcdB fueron amplificados y digeridos con diferentes enzimas de restricción: EcoRI para tcdA; HincII y AccI para tcdB. Además, se evaluó la presencia de cdtB (gen de la toxina binaria B) y de las toxinas A y B (por dot blot), así como el efecto citotóxico de sobrenadantes de cultivo sobre células Vero. En conjunto, estos estudios revelaron tres toxinotipos circulantes según la clasificación de Rupnik: 0, I y VIII; el más prevalente fue el último. Aunque son necesarios más estudios (ej., secuenciación), es interesante notar que la presencia del toxinotipo I podría estar relacionada con la introducción de bacterias de diferente origen geográfico.
En los pacientes infectados con el toxinotipo VIII, el análisis multivariante de los resultados de laboratorio mostró que los aislamientos asociados a decesos (GCD13, GCD14 y GCD22) estaban situados en regiones de los biplots relacionados con valores de laboratorio alterados al momento de la internación. En los otros pacientes, aunque no se observó correlación entre los valores de laboratorio al momento de la internación y la evolución clínica, los niveles de urea, creatinina y recuento de glóbulos blancos estuvieron correlacionados positivamente entre sí una vez diagnosticada la infección.
Nuestro estudio revela la circulación de diferentes toxinotipos de C. difficile en un mismo hospital público. La variedad de toxinotipos puede originarse a partir de microorganismos preexistentes en la región, así como también por la introducción de bacterias provenientes de otras regiones geográficas. La existencia de microorganismos con diferente potencial patogénico es relevante para el control, el seguimiento y el tratamiento de las infecciones.
Clostridioidesdifficile16,24 is a spore-forming, anaerobic pathogen that can be found in the intestinal tract of mammals as well as in public spaces8,32. C. difficile is responsible for 13–30% of antibiotic-associated diarrhea23,24. The pathologic process is principally triggered by two toxins, TcdA (enterotoxin) and TcdB (cytotoxin)11,15. These toxins are codified in a 19.6kb chromosomal region, the pathogenicity locus (PaLoc), harboring tcdA and tcdB and accessory genes17,0,21. Non-toxigenic strains replace PaLoc for a highly conserved 115/75bp non-coding region6.
A third toxin, the binary toxin (CDT), can be produced by C. difficile. It is encoded by two genes, cdtA and cdtB, localized in a 6.2kb chromosomal locus (CdtLoc)20. Although the role of CDT in the pathogenesis of C. difficile has not been univocally established, it is thought that CDT is involved in adherence and colonization of C.difficile26.
Typing methods have been widely used to: (1) evaluate outbreaks9, (2) detect new strains with different pathogenic potential3,10 and (3) gain insight into the spread of C.difficile14.
Toxinotyping of C difficile is based on the variability of the PaLoc region28,29,30. These variations are assessed by PCR- amplification of 5′-end of tcdB (B1) and 3′-end of tcdA (A3), followed by the restriction fragment length polymorphism profile (PCR-RFLP) analysis and digestion with specific enzymes. Currently, these profiles, in addition to the ability to produce TcdA and TcdB, the presence of genes related to the binary toxin (CDT), and the pattern of cytotoxic effects on cell cultures allow for the definition of 34 toxinotypes29.
The aim of the present work was to characterize the circulating toxinotypes of C.difficile in a hospital in Ciudad Autónoma de Buenos Aires, Argentina.
Materials and methodsSamplesStool samples (n=132) were collected between March and September 2016 and analyzed by using the AlereTechLab C.DIFF.QUIK COMPLETE test® or RIDA®QUICK test according to the manufacturer's instructions.
Blood samples from patients with a positive result for C. difficile infection (CDI) were obtained through standard procedures. According to the guidelines for the identification of severe cases of CDI, serum urea (Urea cinética AA, Wiener Laboratorios S.A.I.C, Rosario, Argentina), serum creatinine (Jaffe method; Wiener Laboratorios S.A.I.C, Rosario City, Argentina), and white blood cell counts, were also assessed. Laboratory values at admission (LVA) and after the onset of clinical symptoms (LVOS) were recorded.
Isolation procedureFecal samples were treated with ethanol (1:1) for 30min at room temperature. Then, the material was homogenized with sterile phosphate buffered saline (PBS; 0.144g/l KH2PO4, 9g/l NaCl, 0.795g/l Na2HPO4, pH 7.5). Afterwards, suspensions were streaked on reinforced clostridial medium- (RCM) agar (Laboratorios Britania S.A., Argentina) supplemented with 0.1% (w/v) sodium taurocholate (Santa Cruz Biotechnology, Dallas, TX, USA). Plates were incubated for 48h at 37°C under anaerobic conditions (AnaeroPak; Mitshubishi Gas Chemical Co, Inc.). Colonies were selected based on morphology and Gram staining and genetically characterized as indicated below.
Genetic characterization of C.difficile clinical isolates.
DNA extractionPresumptive C. difficile isolates were grown in BHI broth (BHI: Biokar Diagnostic, Beauvais, France) supplemented with 0.05% (w/v) L-cysteine for 48h at 37°C under anaerobic conditions (AnaeroPak; Mitshubishi Gas Chemical Co, Inc.). After incubation, 1ml of the culture was centrifuged (16000g, 3min) and the pellet was stored at -20°C until use.
Three strains were used as controls: VPI 10463 strain (Ribotype 087) (tcdA+, tcdB+, cdtA−, cdtB−), ALCD3, a clinical isolate (tcdA+, tcdB+, cdtA+, cdtB+) and the non-toxigenic strain ATCC 43593 (Ribotype 060, tcdA−, tcdB−, cdtA−, cdtB−).
After thawing, pellets were washed with 1ml of 0.1M NaCl, suspended in 300μl of 6% (w/v) CHELEX (BIO-RAD, USA) and incubated at 60°C for 20min. Samples were vortexed, incubated at 100°C for 8min, centrifuged at 16000g for 3min, aliquoted and stored at −20°C until use.
Characterization of clinical isolatesTo identify Clostridioides at the genus level, sequence codifying 16S ribosomal RNA (rRNA) of Clostridioides spp. was used38. Presence of the PaLoc region was detected by using Lok3/Lok1 primers, specific for C. difficile species. Strains having the PaLoc region do not show amplification products with these primers. All the studied isolates analyzed for toxinotypes were positive for the PaLoc region, thus indicating that they belong to C. difficile species. For the detection of the cdtB gen, the primer cdtB was used. Details on the primers and PCR conditions are included as supplementary material (Tables S1 and S2).
Reactions were performed using a Taq-polymerase kit (Taq PEGASUS, Productos Bio-Logicos, Argentina) and DNA samples were resolved in 1% w/v agarose gels (Biodynamics).
RFLP analysisAnalysis of restriction fragment length polymorphism (RFLP) was performed according to Rupnik's method30 with some modifications (http://www.mf.um.si/mf/tox/profile.html). The PaLoc region was analyzed by using primers a3c/a4n and b1c/b2n targeting the tcdA and tcdB genes, thus leading to A3 and B1 fragments respectively. Details on the primers and PCR conditions are included as supplementary material (Tables S1 and S2).
Amplified fragments were visualized on 1% (w/v) agarose. PCRs were performed by using a Polymerase Kapa kit (Laboratorios Biolabs S.A., Argentina).
To determine the RFLP pattern, the A3 fragment was digested with EcoRI (Biolabsinc, New England) and the B1 fragment was digested with HincII and AccI (Biolabsinc) according to the manufacturer's instructions. Both fragments, A3 and B1, were digested at 37°C for 30min. Both digested and non-digested samples were analyzed by electrophoresis on 1.5% (w/v) agarose gels25.
Cell culturesVero cells were grown in Dulbecco's Modified Eagle's Medium (DMEM; Gibco BRL Life Technologies, Rockville, MD, USA) supplemented with 10% v/v inactivated (30min/60°C) fetal calf serum (Natocor Industrias Biológicas, Córdoba, Argentina), 2g/l NaHCO3, 10mg/l streptomycin and 10IU/ml penicillin G and 1% (v/v) non-essential amino acids (Life Technologies). Cells were seeded at 75000 cells/well in 96-well tissue culture plates (JetBiofil®, Guangzhou, China) and incubated at 37°C for 48h in a 5% (v/v) CO2–95% (v/v) air atmosphere.
Cytotoxicity assay on Vero cellsBacterial isolates were grown in BHI broth and centrifuged as indicated above. Supernatants were filter-sterilized (0.45μm pore diameter) and stored at -80°C until use. Before the cytotoxicity assay, Vero cells were washed twice with PBS. Spent culture supernatants (SCS) were serially (two-fold) diluted in DMEM without fetal calf serum. One hundred microliters of diluted SCS were added per well and incubated at 37°C for 16h in a 5% (v/v) CO2/95% (v/v) air atmosphere. Cell rounding and morphological changes were evaluated by phase contrast microscopy36. Type of cytopathic effect was analyzed according to Rupnik et al.27.
Detection of A and B toxins by dot blotPresence of A and B toxins in SCS was assessed by the dot blot assay. Briefly, 4μl of SCS were spotted on a nitrocellulose membrane. Blocking was performed with 3% (w/v) skim milk-TTBS (50mM Base Trizma (Hydroxymethyl aminomethane Mallinckrodt, Baker Inc.), 150mM NaCl and 0.05% (w/v) Tween 20 (Sigma–Aldrich, Inc., St. Louis, MO, USA), pH 7.5 for 1h at 37°C. Membranes were incubated for 40min at 37°C with mouse anti-TcdA (1/1000) or anti-TcdB (1/500) monoclonal antibodies (Meridian Life Science Inc., USA). Next, membranes were incubated with 1/1000 biotinylated mouse anti-IgG (Sigma–Aldrich, Inc., St. Louis, MO, USA) for 30min at 37°C. All dilutions were made in 1% w/v skim milk-TTBS. After streptavidin alkaline phosphatase (BD Pharmingen, USA) was added, membranes were incubated for 30min at 37°C and revealed with NBT/BCIP commercial substrate (Aldrich, Inc., St. Louis, MO, USA) according to the manufacturers instructions.
Statistical analysisMultivariate analysis was conducted by using Infostat Software (InfoStat versión 2013. Grupo InfoStat, FCA, Universidad Nacional de Córdoba, Argentina).
Clinical isolates belonging to the most prevalent toxinotype (VIII) were further analyzed in the context of laboratory data from individuals harboring these strains. This multivariate analysis included laboratory data (creatinine, urea and white blood cell counts) as variables.
In biplots, vector variables represent the positive direction of the variable axes. Lengths of these vectors approximate the standard deviation of the variables. The angle between two variable vectors approximates the arc cosine of the correlation between those variables. Therefore, variables forming an acute angle are positively correlated, whereas those forming an obtuse angle are negatively correlated. Right angles indicate uncorrelated variables. Each isolate is denoted by a circle whose coordinates correspond to the principal component scores.
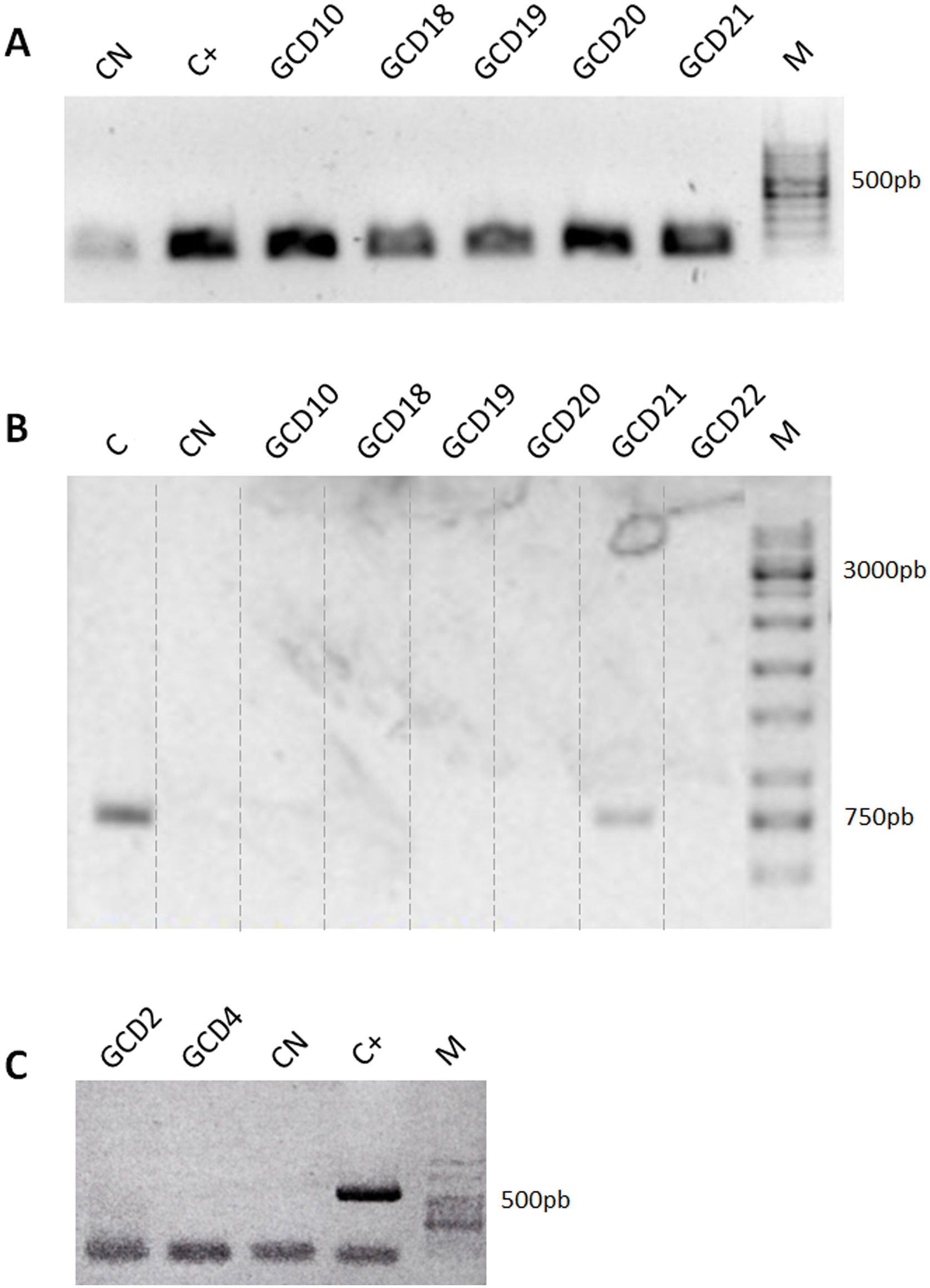
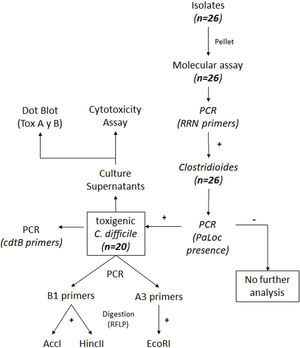
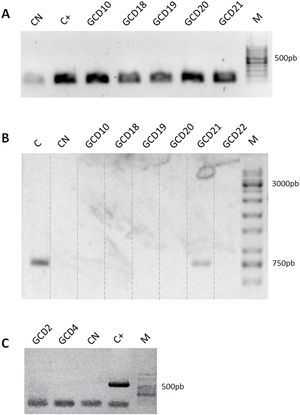
ResultsIsolation of C. difficile strains and determination of toxin-associated genesDetails on the steps followed for the isolation and identification of C. difficile samples are shown in Figure 1. Twenty-six isolates compatible with C. difficile were recovered from the samples analyzed. Each isolate comes from a different sample and samples were from different patients. All 26 isolates were positive for the rrn sequence (100bp) specific to the genus Clostridioides (Fig. 2A).
Detection of Clostridioides-associated genes. (A) Determination of rrn amplicons specific for genus Clostridioides. Positive control (C+): C. difficile ALCD3; negative control (CN): water. (B) Determination of Tox- amplicons to determine the presence of the PaLoc island. Positive control (C): C. difficile ATCC 43593 (non-toxigenic strain); negative control (CN): water. (C) Detection of binary toxin cdtB gene. Positive control (C+): C. difficile ALCD3; negative control (CN): water. M: molecular weight size marker.
In order to detect the presence of the pathogenicity island (PaLoc), primers Lok3 and Lok1 were used. These primers were located outside the PaLoc region, thus, the absence of amplification with those primers indicates that the PaLoc region is present. On the contrary, if amplification with Lok3 and Lok1 primers occurred, the isolate is considered non-toxigenic. Results showed that 20 out of 26 isolates analyzed were toxigenic. As an example, both patterns are shown in Figure 2B: toxigenic isolates (GCD10, 18, 19, 20 and 22) and a non-toxigenic isolate (GCD21). In addition, all isolates were negative for the binary toxin gene (cdtB) (Fig. 2C).
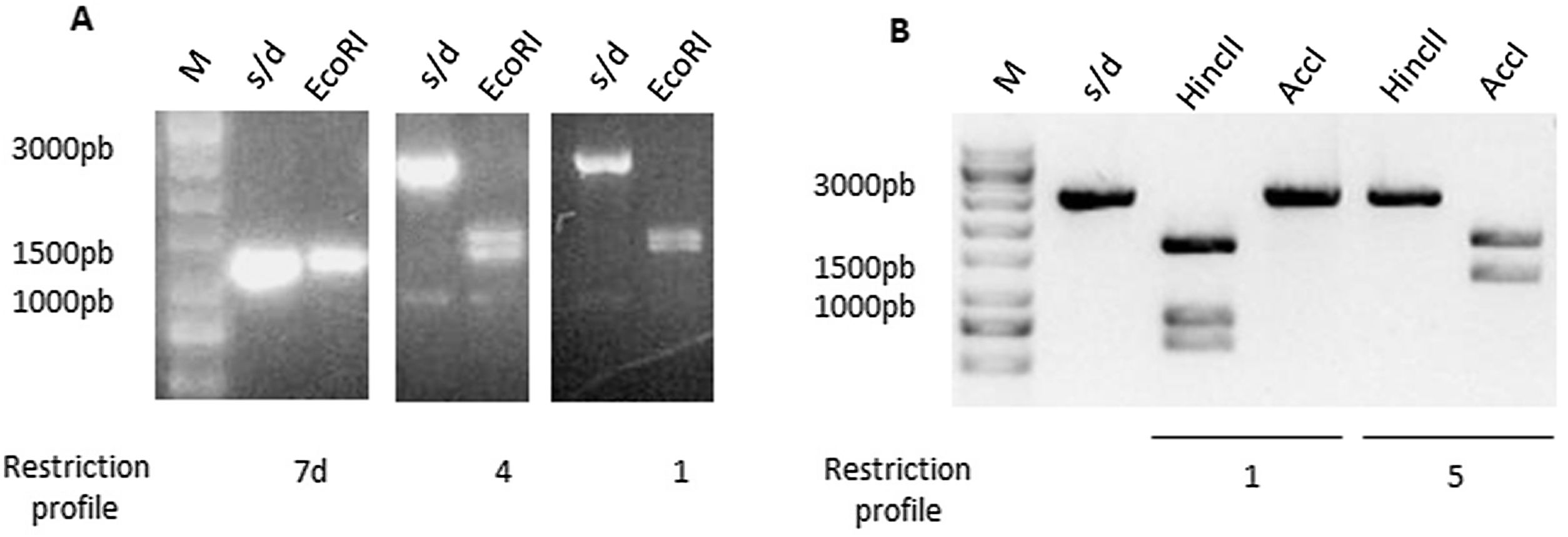
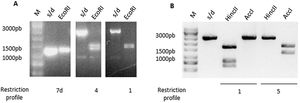
ToxinotypingDetermination of the restriction fragment length polymorphism pattern (RFLP)Different toxinotypes of C.difficile arise from polymorphisms in the PaLoc island and could lead to different virulence levels13. For toxinotyping, tcdA (A3 fragment) and tcdB (B1 fragment) genes were amplified with primers a3c/a4n and b1c/b2c, respectively. EcoRI was used to digest the A3 fragment whereas AccI and HincII restriction enzymes were used separately to digest the B1 fragment. Digestion of the A3 fragment led to restriction profiles 1, 4 and 7d, whereas the digestion of the B1 fragment led to profiles 1 and 5. Representative digestion profiles are shown in Figure 3.
Restriction Fragment Length Polymorphism (RFLP) of TcdA and TcdB amplicons. (A) TcdA fragment digested by EcoRI showed three different restriction profiles: 7d, 4 and 1. (B) TcdB fragment digested by HincII and AccI showed two different restriction profiles: 1 and 5. M: molecular weight size marker.
The combination of A3 and B1 profiles allowed us to identify 3 different toxinotypes. Two isolates were associated with toxinotype 0 (GCD4 and GCD27); 1 isolate with toxinotype I (GCD18) and 17 with toxinotype VIII (GCD2, GCD3, GCD10, GCD13, GCD14, GCD15, GCD16, GCD17, GCD19, GCD20, GCD22, GCD23, GCD24, GCD25, GCD26, GCD28 and GCD29) (Table 1).
Characterization of C. difficile isolates.
| Toxinotype | Isolate or strain | Dot Blota | CdtB PCRb | RFLPc | Type of CPEd | ||
|---|---|---|---|---|---|---|---|
| TcdA | TcdB | B1 (HincII+AccI) | A3 (EcoRI) | ||||
| 0 | VPI 10463e | + | + | − | 1 | 1 | D |
| GCD4 | |||||||
| GCD27 | |||||||
| 0/v | ALCD3e | + | + | + | 1 | 1 | D |
| I | GCD18 | + | + | − | 1 | 4 | D |
| VIII | GCD2 and 3 GCD10GCD13 to 17 GCD19 to 20 GCD22 to 26 GCD28 to 29 | − | + | − | 5 | 7d | S |
CPE: cytopathic effect; D: difficile-like effect; S: sordellii-like effect.
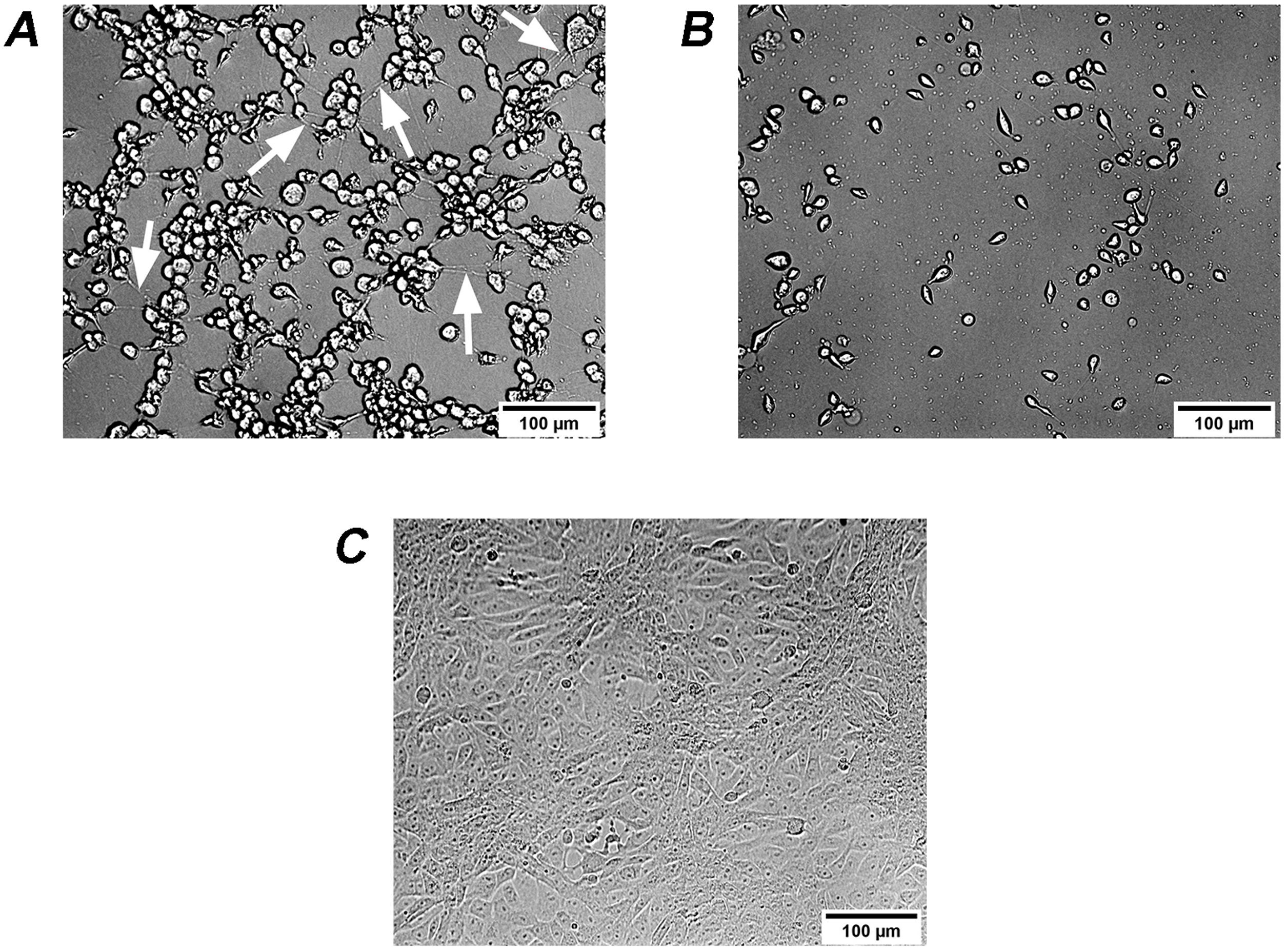
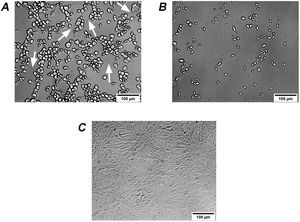
It is known that the biological activity of TcdB is 100–10000 times higher than that of TcdA35. Therefore, the effects on Vero cells of extracellular factors from C. difficile are mainly associated with TcdB2,5. Different isoforms of TcdB lead to changes in the activity and/or recognition specificity to Rho proteins, thus leading to differences in cytopathic effects13,22. Therefore, two types of cytopathic effects on Vero cells, i.e. cell rounding with long protrusions (difficile-type damage or D-damage) and complete cell rounding without protrusions (sordellii-type damage or S-damage)27, have been described. Analyzed culture supernatants from 3 isolates led to difficile-type damage on Vero cells: GCD4, GCD18 and GCD27 (Fig. 4A), while the remaining 17 isolates led to sordellii-type damage on Vero cells (Fig. 4B).
Cytopathic effect (CPE) of culture supernatants on Vero cells. (A) GCD4 supernatant induces cell rounding with remaining long protrusions (white arrows) called difficile-type damage or D-damage; (B) GCD10 supernatant induces complete cell rounding form called sordellii-type damage or S-damage; (C) control cells.
The presence of TcdA and TcdB in culture supernatants of C. difficile was detected by dot blot using monoclonal antibodies (a-TcdA or a-TcdB). Both toxins were detected in culture supernatants from GCD4, GCD18 and GCD27 isolates. Strain VPI 10463 was used as positive control. The remaining 17 C. difficile isolates were positive for TcdB but negative for TcdA (Table 1).
Results from binary toxins, RFLP, biological assays and the detection of TcdA and TcdB toxins were analyzed in order to determine the toxinotype for each isolate as well as the toxinotypes of reference strain VPI 10463 and isolate ALCD3. As shown in Table 1, toxinotype VIII was the most prevalent one (17 isolates), followed by toxinotype 0 (2 isolates) and toxinotype I (one isolate).
Multivariate analysisThe isolates belonging to the most prevalent toxinotype (VIII) were selected for multivariate analysis considering laboratory data as variables. Figure 5 shows that the principal components CP1, CP2 and CP3 explain 90% of the variation in the dataset. The two-dimensional scatter diagram constitutes a good approximation to the original dataset of a six-dimensional space (one dimension for each variable studied). As shown in Figure 5, CP1 is related to increased values in white blood cells (WBC), creatinine and urea after the onset of symptoms. Noteworthy, isolates GCD14, GCD25 and GCD28 are associated with high white blood cell counts as well as high levels of urea and creatinine after the onset of symptoms. Interestingly, strains associated to fatal outcomes (GCD13, GCD14 and GCD22) can be found in regions related to altered laboratory values at admission. Concerning the correlation between laboratory values, as expected, after C. difficile infection was diagnosed (LVOS), the levels of urea, creatinine, and white blood cells were positively correlated. In contrast, variables corresponding to laboratory values at admission (LVA) were not correlated.
Multivariate (principal component analysis) of laboratory data from individuals with C. difficile diagnosis. Each value represents a C. difficile isolate. Samples were analyzed at patient admission to hospital and after the onset of clinical symptoms of C. difficile infection. The percentages of the variation explained by principal components (CP1, CP2 and CP3) are indicated in parentheses. References: LVA: Laboratory Values at Admission and LVOS: Laboratory Values after the Onset of Symptoms.
Since the beginning of the 21st century the incidence and severity of CDI has increased worldwide. Changes in epidemic dynamics are principally associated with the emergence of hypervirulent strains, i.e. ribotypes 027, 078 and 2448. To control the spread of CDI, epidemiological studies are necessary to assess the distribution of circulating strains as well as their pathogenic potential1.
The analysis of the isolates based on the presence of sequences of genes tcdA, tcdB and cdtB, revealed that all the isolates belong to genotype tcdA+/tcdB+/cdtB−. However, the results obtained by immunoblots showed two patterns: TcdA+/TcdB+ and TcdA−/TcdB+.
Toxinotyping of C. difficile isolates by RFLP assess genetic variations of the PaLoc island that give toxins with different biological activity as well as different interaction with antibodies25,28,29. This variability is relevant for clinical and diagnostic purposes.
Strain VPI 10463 was used as reference strain (Toxinotype 0). Other different toxinotypes show differences in the PaLoc island when they are compared with the reference strain29. These differences are due to polymorphisms or deletions in this region. For example, toxinotype I (a minor toxinotype) exhibits deletions or RFLPs in the tcdA gene, and toxinotype VIII exhibits RFLPs in the tcdB gene and a 1.8 Kb deletion at the 3′end of the tcdA gene coding for the c-terminal portion of TcdA (indeed this is a TcdA(−) toxinotype)34. In the present work, when the RFLP analysis was conducted, 3 toxinotypes were detected: 0, I and VIII (prevalence 10%, 5% and 85%, respectively). These results are in agreement with previous reports showing that toxinotypes III, IV, V and VIII represent the most common toxinotypes present in isolates from human origin27.
Interestingly, strains belonging to toxinotype VIII show variability in the catalytic domain of TcdB, leading to a homologous amino acid sequence of the Lethal Toxin of Clostridum sordelli (LTCS)7. As a consequence, both TcdB variant and LTCS glycosylate similar R-Ras substrates give rise to a cytopathic effect characterized by cell rounding without protrusions (Sordelli-like cytopathic effect).
It is known that some toxinotypes (e.g. III and VIII) are related to increased virulence and relapses28. Interestingly, toxinotype VIII, frequently isolated from asymptomatic infants16, was reported as being responsible for outbreaks in England, The Netherlands, Poland and Ireland7. In Argentina, nosocomial strains have been characterized and toxinotype VIII has also been reported4,12,37.
It is worth noting that isolate GCD18 belongs to the minor toxinotype I, characterized by a deletion and RFLPs in the tcdA gene. Even though more studies are needed (e.g. sequencing analysis), the occurrence of this toxinotype in the hospital is compatible with the introduction of bacteria from different sources, because it is known that this toxinotype arises from the recombination of CROP regions situated in tcdA29,39. This fragment is lacking in toxinotype VIII, the most prevalent toxinotype detected in the hospital studied. Most of the strains of a given ribotype have similar sequences in the PaLoc region, thus belonging to a single toxinotype26. In contrast, a single toxinotype includes several ribotypes31. Noteworthy, one of the toxinotypes found in the present study, toxinotype VIII, is compatible with ribotypes 017, 047 and 11026. The most probable ribotypes for isolate GCD18 (toxinotype I) are 003; 012 and 10228. From the data obtained in the medical records (data not shown), we observed that the patients infected with GCD13, GCD14, GCD22 isolates exhibited SOFA (Sequential Organ Failure Assessment) scores33 above 2 points. Those isolates were associated to fatal outcomes. Patients infected with GCD14 and GCD22 isolates presented pulmonary tuberculosis as associated comorbidity. Those patients infected with GCD24, GCD25 and GCD28 isolates were associated with renal failure secondary to a greater number of diarrheal stools. Patients infected with GCD15 and GCD28 isolates were admitted with renal failure but progressed with clinical and chemically improved symptoms after treatment.
Isolates were from a hospital which receives patients from different regions of the center of Buenos Aires Province. Although recommended practices for the prevention of healthcare-associated infections are implemented, the control of the circulation of sporulated microorganisms is a challenging issue. Noteworthy, in addition to the toxinotypes reported here, other toxinotypes (e.g. 0/v and III) were also circulating in other hospitals of the province of Buenos Aires, Argentina (data not shown). As expected, the most prevalent toxinotype (VIII) was always present. This finding is in agreement with results reported by Quemeneur et al.25.
Although reports on the characterization of circulating strains of C. difficile in Argentina are still scarce, there are studies on circulating strains performed by different methodological approaches and there are reports on the circulation of TcdA(−), TcdB(+) as well as CDT (+) and the epidemic strain ST11,4,12,18,19.
The potential worldwide spread of CDI calls for epidemiological studies to characterize currently circulating strains and highlights the need for increasing surveillance. The results presented here could contribute to gain further insight into the pathogenesis of C. difficile as well as to delineate control strategies.
Conflicts of interestNone.
Ethical responsibilitiesNone.
The present work is part of the legacy of Dr. Eduardo Alul. His motivation, help and encouragement were certainly paramount driving forces for this collaborative study.
Authors acknowledge financial support from Facultad de Ciencias Exactas (Universidad Nacional de La Plata), CONICET and ANPCyT. All the isolates were from HIGA Luisa G de Gandulfo Hospital, Lomas de Zamora, Buenos Aires, Argentina. Strain ALCD3 was kindly provided by Liliana Fernández Canigia from the Microbiology Laboratory of the Hospital Alemán (Buenos Aires, Argentina).