Salmonellaenterica serovar Enteritidis (S. Enteritidis) is the most frequent serovar involved in human salmonellosis. It has been demonstrated that about 80% of infections are related to biofilm formation. There is scant information about the pathogenicity of S. Enteritidis and its relationship to biofilm production. In this regard, this study aimed to investigate the differential host response induced by S. Enteritidis biofilm and planktonic lifestyle. To this purpose, biofilm and planktonic bacteria were inoculated to BALB/c mice and epithelial cell culture. Survival studies revealed that biofilm is less virulent than planktonic cells. Reduced signs of intestinal inflammation and lower bacterial translocation were observed in animals inoculated with Salmonella biofilm compared to the planktonic group. Results showed that Salmonella biofilm was impaired for invasion of non-phagocytic cells and induces a lower inflammatory response in vivo and in vitro compared to that of planktonic bacteria. Taken together, the outcome of Salmonella–host interaction varies depending on the bacterial lifestyle.
Salmonella enterica serovar Enteritidis (S. Enteritidis) es la serovariedad más frecuentemente aislada en la salmonelosis humana. Se ha demostrado que alrededor del 80% de las infecciones están relacionadas con la formación de biopelículas. Sin embargo, la información disponible acerca de la patogenicidad de S. Enteritidis y su relación con la producción de biopelículas es escasa. Este trabajo tuvo como objetivo investigar la respuesta diferencial del huésped frente a S. Enteritidis en sus 2 estilos de vida: biopelícula y planctónico. Para ello, se inocularon bacterias en estado de biopelícula o planctónico en ratones BALB/c y cultivo de células epiteliales. Los estudios de supervivencia revelaron que Salmonella en biopelícula fue menos virulenta que su contraparte planctónica. Los animales inoculados con biopelículas presentaron una mayor conservación estructural del intestino y una menor translocación bacteriana que el grupo planctónico. Asimismo, Salmonella en biopelícula mostró una capacidad deficiente para invadir células no fagocíticas e indujo una menor respuesta inflamatoria in vivo e in vitro que las bacterias planctónicas. Se concluye que el resultado de la interacción Salmonella-huésped depende del estilo de vida bacteriano.
Salmonellosis is one of the most important foodborne illnesses worldwide, including Argentina35. The Global Burden of Diseases, Injuries, and Risk Factors Study (GBD) estimated that Salmonella enterocolitis occasioned 95.1 million cases and 50,771 deceases in 201710. Salmonellaenterica serovar Enteritidis (S. Enteritidis) is the most frequent foodborne bacteria associated with the consumption of egg products and undercooked poultry29 and about 80% of infections are related to biofilm formation6. Several studies demonstrated that different sets of genes are expressed in bacterial biofilms compared with planktonic cells25,28,32. The most important constituents of the matrix of Salmonella biofilms are cellulose and curli amyloid fibril structures. There are other components such as the O antigen, extracellular DNA, fatty acids, and surface proteins such as BapA33. The transcriptional regulator CsgD activates the switch from the planktonic lifestyle to biofilm formation30. As a master biofilm regulator, among other functions, CsgD positively regulates the expression of the csgBA operon involved in curli fibril production5. Furthermore, CsgD acts on the promoter region of adrA, which controls the synthesis of c-di-GMP activating at a posttranscriptional level the cellulose synthase from the bcs operon34.
Traditionally the ability of Salmonella to persist under stressful conditions separately from pathogenesis has been studied. Nevertheless, it is clear that there must be an intimate relationship between both traits considering its continuous success as a pathogen17. Although many of the pathogenesis mechanisms of Salmonella are described as a result of host-bacterial studies, most of these investigations were developed with planktonic bacteria. In this regard, this study aimed to investigate the differential host response induced by S. Enteritidis biofilm and planktonic lifestyle.
Materials and methodsAnimal ethics statementEight to 10-week old female BALB/c mice were used. Animals were maintained in our vivarium under standard conditions. Water and food were provided ad libitum. Mice were sacrificed with carbon dioxide followed by cervical dislocation. All procedures were approved by the Institutional Animal Care and Use Committee (CICUAL), School of Medicine, University of Buenos Aires (approval number 50953/2016). The statistical analysis justified the number of mice required and was approved by CICUAL.
Bacterial strain and growth conditionsS. Enteritidis 5694 wild type strain isolated from human stool was kindly provided by Dr. Anne Morris Hooke, Miami University. The strain originally belonged to Dr. F. Collins’ collection, Trudeau Institute, Saranac Lake, New York8. Biofilm and planktonic-cells were obtained from the same flask incubated under biofilm-inducing conditions as follows: S. Enteritidis 5694 was plated on trypticase soy agar (TSA, Britania S.A, Argentina) and incubated at 37°C for 20h. One colony was then transferred to 10ml of trypticase soy broth (TSB, Britania S.A, Argentina) and was grown at 37°C and 200rpm until optical density OD590 0.6. Then, 3ml of the bacterial culture were centrifuged (8000×g; 5min; 4°C), washed and suspended in 1ml of Luria-Bertani broth (LB) without salt, pH 7.0 (LBNaCl-). This suspension was diluted to OD590 ∼0.05 in 240ml of LBNaCl- and distributed in 40ml volumes into six separated tubes containing a sterile glass coupon (75mm×25mm) immersed in the media. Tubes were incubated at 28°C without agitation for 48h. The biofilm lifestyle sample was obtained by recovering five coupons with sterile tweezers and non-adherent bacteria were eliminated by two successive soakings in physiological solution (PS). The biofilm grown on both side surfaces of the coupons was recovered and collected in a 1.5ml tube using a scratcher and 1ml of PS. Cells were then dispersed by vortexing and homogenized by passage through a syringe with a 22 gauge needle ten times. The proper biofilm disaggregation was confirmed by observing only single cells with no bacterial aggregates using optical microscopy. The sixth culture tube was used as a control for proper biofilm production on glass coupons by fluorescence microscopy (Eclipse E600, Nikon, USA) using calcofluor (Fluorescent Brigthener 28, Sigma-Aldrich, Argentina) for cellulose staining as described in Aya Castañeda et al.4 The planktonic lifestyle sample was obtained by collecting 1ml of fluid phase from each of the five tubes. To get rid of multicellular aggregates, this suspension (5ml) was centrifuged at low speed (200×g; 2min; 4°C) and the supernatant was transferred into a new Falcon tube and centrifuged at high speed (8000×g; 5min; 4°C) and the pellet was homogenized by vortexing in PS. Bacterial cultures were standardized so that the results of the CFU/inoculum were reproducible. The number of bacteria was determined for both lifestyle samples before animals and HEp-2 (ATCC CCL-23, Manassas, USA), inoculation by counting colony forming units (CFU) after plating appropriate dilutions of each sample on TSA plates incubating them for 24h at 37°C.
Bacterial RNA extraction and reverse transcriptionBacteria were grown as described above. Total RNA extraction was performed using TRIzol reagent (Invitrogen, California, USA) as described in Noto Llana et al.22 The expression of csgD, bcsA, csgA, adrA, bapA and 16S (endogenous normalizer) genes was analyzed by real-time quantitative polymerase chain reaction (qPCR) as described below. Primer sequences are shown in Suppl. Table S1.
Mice inoculation, survival assay and clinical scoreAnimals were randomly divided into five groups (eight mice per group) according to the treatment they received: planktonic cells, biofilm or control. Salmonella intragastric inoculation was performed as described in Giacomodonato et al.13 Two different bacterial doses were used, 103 and 104 CFU per animal. For the survival assay, animals were monitored daily for weight and clinical conditions during ten days post-inoculation. Animals with >20% weight loss were euthanized. Clinical condition was evaluated using the score scale established by Alam et al.3 Clinical scoring was performed following a double-blinded protocol. The scores of individual animals in one group were compared with those of individual animals in the other group to calculate statistical significance.
Splenic bacterial colonization, RNA extraction and reverse transcriptionAnimals were randomly divided into five groups (five animals per group) according to the treatment they received: planktonic cells, biofilm or control and were inoculated intragastrically with two different bacterial doses, 103 and 104 CFU, per animal as indicated above. Mice were euthanized at day 5 post-inoculation. Spleen and intestinal samples (ileum) were harvested aseptically and immediately processed as described below. Spleens were cut in half and each portion was weighed. For bacterial colonization, one half of the spleen was homogenized in sterile PS, diluted appropriately and cultured on Salmonella-Shigella (SS) agar (Britania S.A, Argentina). After incubation at 37°C for 24h, log CFU/g of tissue was calculated. Negative SS agar cultures were incubated in Selenite broth for 18h. The other half of the spleen was homogenized in TRIzol reagent. Total RNA extraction and RT-PCR were performed as described in Noto Llana et al.22tnf-α, Il-6, Il-17, Il-1β, cox-2 and 18S rRNA (endogenous normalizer) gene expression was determined by qPCR as described below. The primers used are shown in Suppl. Table S1.
Histological analysisIleum samples were extracted from animals as described above (eight mice per group) and processed by histological techniques as described in Noto Llana et al.21 Intestinal histological changes were evaluated by a blinded qualified pathologist using the score scale previously established in our laboratory22.
Murine cytokine analysis by ELISABlood was collected via cardiac puncture 5 days post-infection. TNF-α and IL-6 serum determinations were performed using the following commercial ELISA kits: TNFα (R&D Systems, Minneapolis, USA) and IL-6 (BD OptEIA, BD Biosciences Pharmingen, San Diego, USA) according to the manufacturer's instructions.
Cell culture infection model, RNA extraction and reverse transcriptionConfluent human laryngeal epithelial (HEp-2) cells were infected with Salmonella biofilm or planktonic cells (multiplicity of infection: 10:1) according to Giacomodonato et al.12 At 90min post-infection, monolayers were processed as described in Giacomodonato et al.12 For eukaryotic RNA isolation, the RNeasy Mini kit (Qiagen GmbH, Hilden, Germany) was used following the manufacturer's instructions. cDNA was synthetized using the QuantiTect reverse transcription kit (Qiagen GmbH, Hilden, Germany). The expression of Il-6, Il-8, Il-10 and gapdh (endogenous normalizer) genes was determined by qPCR as described below. The primers used are depicted in Table S1.
Real-time quantitative polymerase-chain-reaction (qPCR)qPCR reaction was performed according to Noto Llana et al.22 Primers are listed in Table S1. PCR parameters were 50°C for 2min, 95°C for 10min, and 40 cycles 15s at 95°C and 30s at 60°C. The relative quantification of gene expression was calculated with the average CT number of each triplicate according to the ΔΔCT method15.
Statistical analysisData were analyzed for statistical significance using a parametric (Student's t test and ANOVA) and a nonparametric test (Mann–Whitney test). Statistical analysis was performed using the GraphPad Prism version 8.0 software program (GraphPad Software, San Diego, CA, USA).
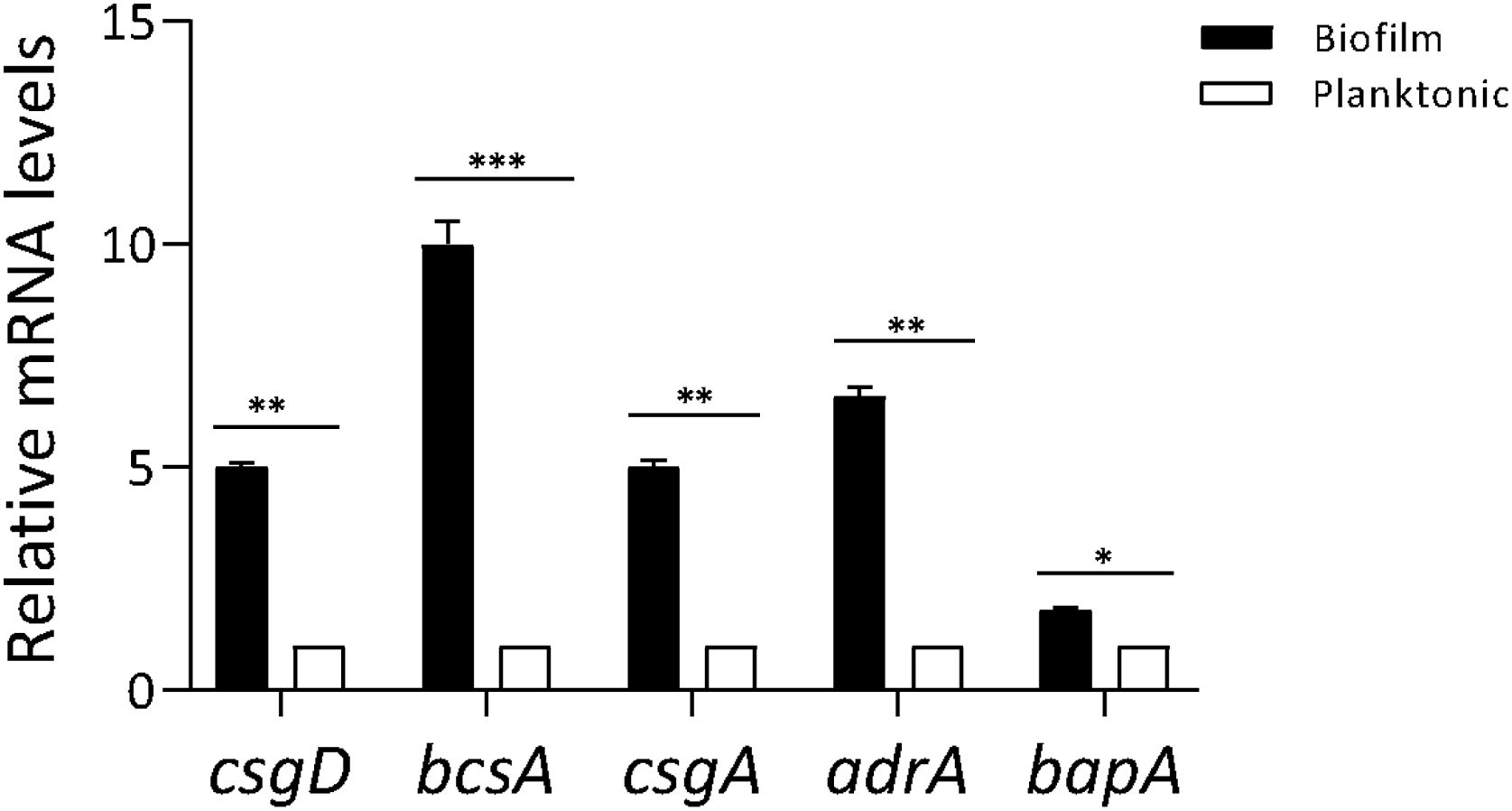
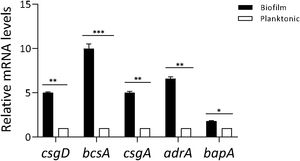
ResultsProfiling gene expression in biofilm and planktonic bacteriaProfiling gene expression in biofilm and planktonic inoculum was determined by qPCR (Fig. 1). The expression of csgD in Salmonella biofilm was significantly increased (p<0.01) compared with their planktonic counterpart. As expected, the expression of both genes bcsA and csgA was significantly increased (p<0.001 and p<0.01, respectively), in biofilm bacteria compared with the planktonic state. Additionally, the expression of other genes associated with biofilm production (adrA and bapA) was determined. As shown in Figure 1, adrA and bapA expression was significantly augmented (p<0.01 and p<0.05, respectively), in the biofilm state compared with the planktonic state.
Profiling gene expression in biofilm and planktonic bacteria. Gene expression in Salmonella biofilm and planktonic inoculum was analyzed by qPCR. The relative mRNA amount was related to that in the planktonic group, set as 1. Results are expressed as mean±SD. Data from three independent experiments were analyzed using the Student t-test. *p<0.05, **p<0.01, ***p<0.001.
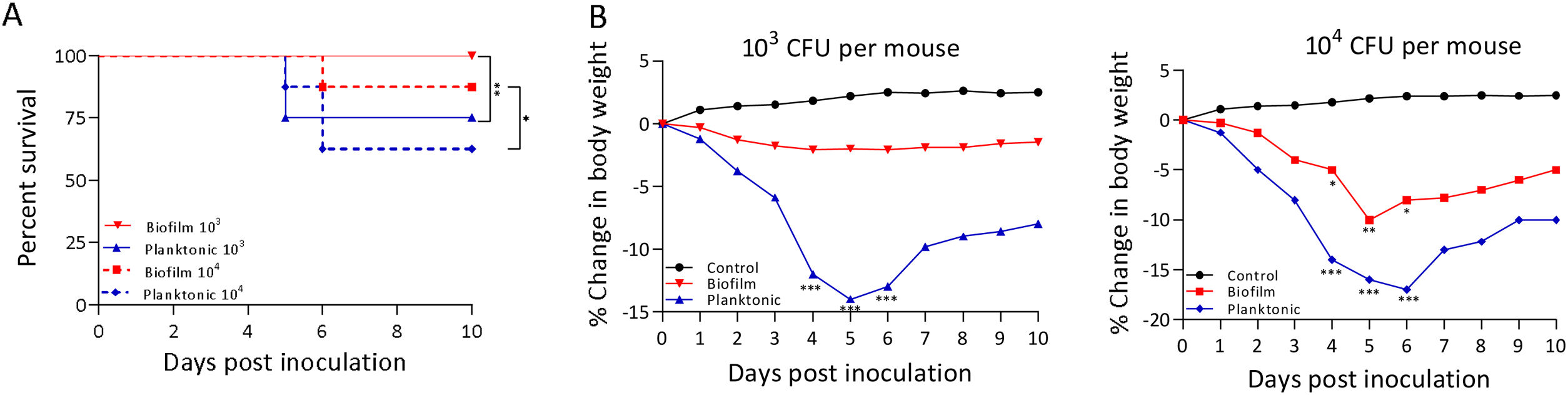
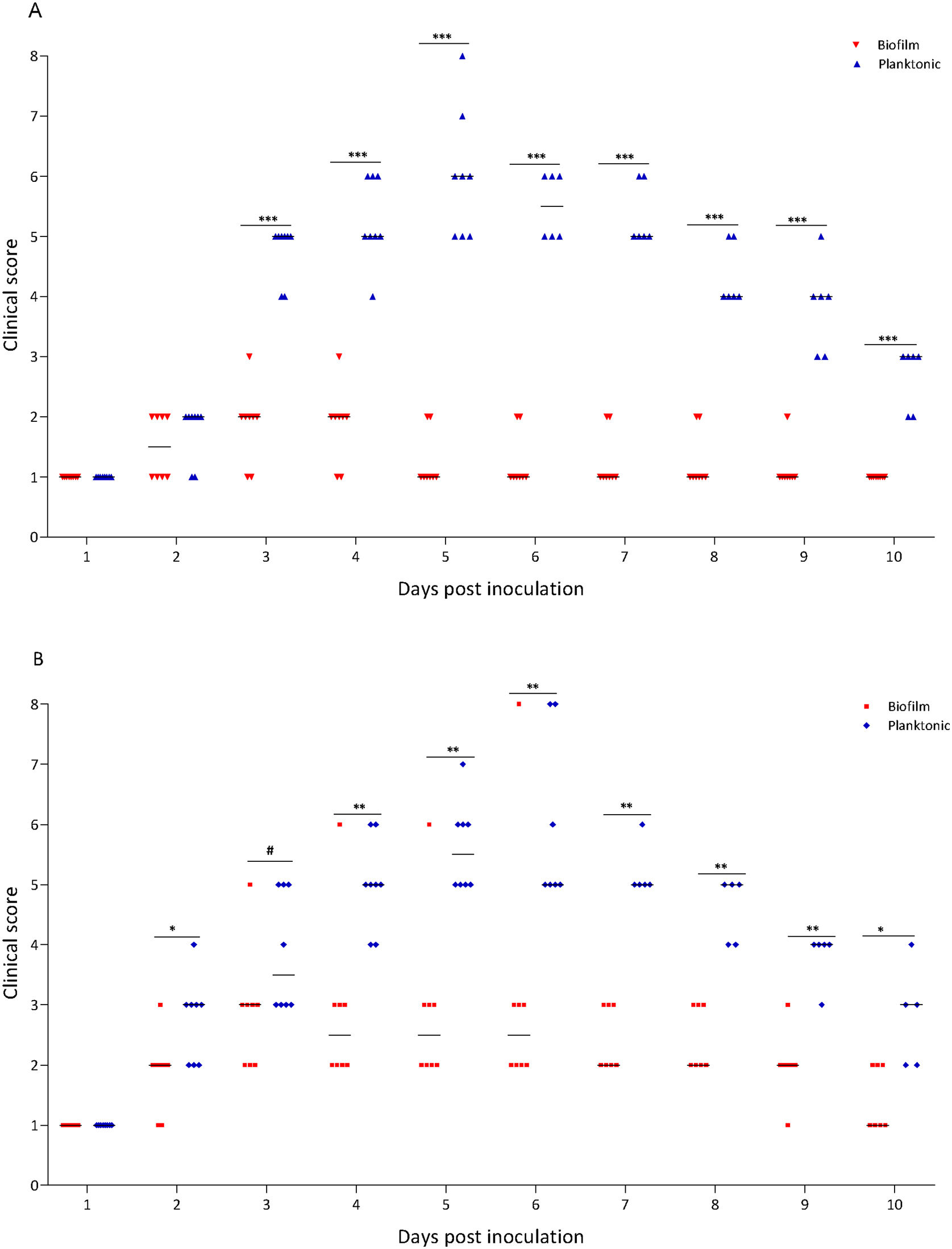
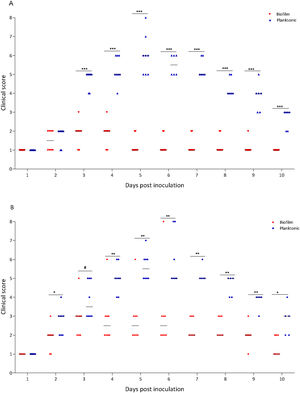
A murine infection model was used to determine whether the biofilm lifestyle alters the virulence characteristics of S. Enteritidis in vivo. To this purpose, BALB/c mice were intragastrically inoculated with two different doses for S. Enteritidis biofilm and planktonic lifestyle. All mice infected with 103 CFU of Salmonella biofilm survived, whereas 2 out of 8 mice (25%) inoculated with the same dose of planktonic Salmonella died at day 5 post-inoculation (p<0.01). Even at higher infective doses the Salmonella biofilm was significantly less virulent than planktonic bacteria. For example, with 104 CFU per mouse, only 1 out of 8 mice (12.5%) in the biofilm group died, whereas 3 out of 8 animals (37.5%) were killed by the planktonic inoculum (p<0.05) (Fig. 2A). Body weight was also measured daily as a general indicator of the overall health of each animal. The percentage change in weights of each group of animals is shown in Figure 2B. Mice inoculated with planktonic Salmonella suffered significant weight loss within a few days compared to the uninfected controls (p<0.001 on days 4, 5 and 6 post-inoculation) (Fig. 2B). Biofilm cells attenuate weight loss in 103 CFU S. Enteritidis-infected mice. On the contrary, animals inoculated with 104 CFU of biofilm bacteria lost significant weight on days 4 (p<0.05), 5 (p<0.01) and 6 (p<0.05) post-infection compared to control animals. Nevertheless, weight loss in mice infected with biofilm bacteria was lower than in those inoculated with planktonic cells. Control mice showed normal weight gain. Furthermore, mice clinical conditions were assessed every day. As shown in Figure 3, from day 2–3 onwards, the differences in clinical score between the two animal groups were statistically significant, both for 103 and for 104 CFU per mouse.
Virulence characteristics of S. Enteritidis biofilm and planktonic lifestyle. Groups of eight mice were inoculated intragastrically with 103 or 104 CFU of Salmonella biofilm or planktonic lifestyle per mouse and (A) survival rate was followed for 10 days. (B) Animals were weighed daily and the percent change in weight from 0 to 10 days is shown. Data were analyzed using the Fisher's exact test. *p<0.05, **p<0.01, ***p<0.001.
Clinical score of mice receiving S. Enteritidis biofilm or planktonic bacteria. Groups of eight animals were inoculated intragastrically with two different doses of Salmonella biofilm or planktonic lifestyle: (A) 103 and (B) 104 CFU per mouse. Clinical conditions were assessed for 10 days, following a double-blinded protocol, and clinical scores were calculated as described in “Materials and methods” section. #p<0.1; *p<0.05; **p<0.01; ***p<0.001. Mann–Whitney U test.
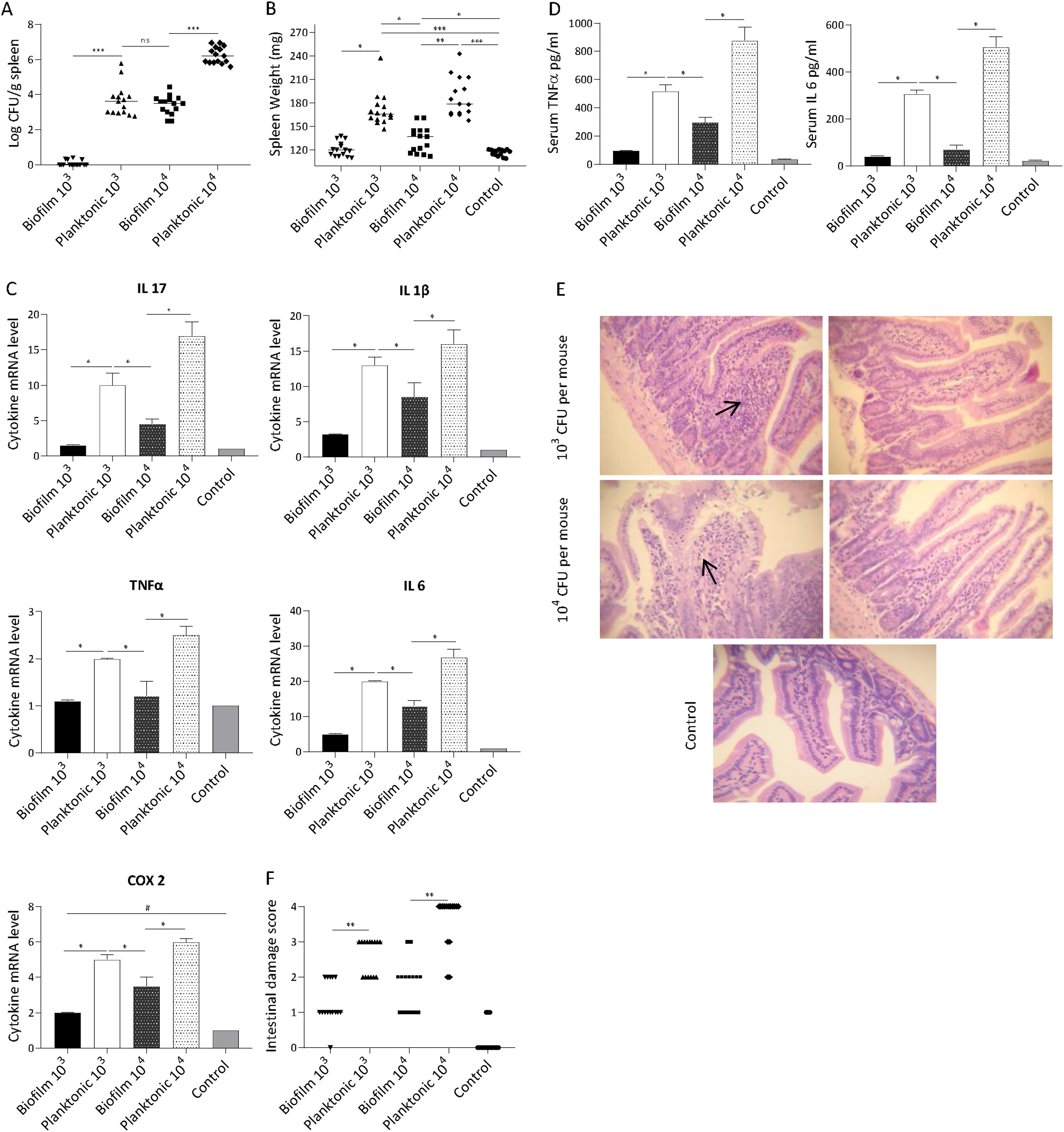
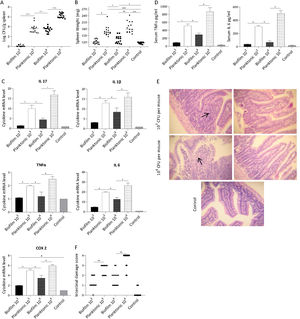
The ability of Salmonella to translocate from the intestine and cause systemic infection is well described31. Thus, we sought to determine if the biofilm lifestyle affects the ability of bacteria to disseminate to other organs. It was found that mice receiving the planktonic inoculum exhibited a significantly higher Salmonella spleen colonization compared with biofilm-inoculated animals. As shown in Figure 4A, at day 5 post-infection practically no Salmonella was recovered from the spleen of animals inoculated with 103 CFU of biofilm bacteria. Concurrently, spleens of mice infected with the same dose of planktonic cells were colonized (median log CFU/g spleen: 3.65). At higher infective doses (104 CFU per mouse) spleens from both animal groups (planktonic and biofilm inoculum) were infected. Nevertheless, bacterial loads recovered from spleens infected with biofilm bacteria were lower (p<0.001) than those found in mice infected with planktonic cells (median log CFU/g spleen: 3.54 vs 6.19, respectively). These results were consistent with a significant difference in spleen weight between biofilm and planktonic cell-inoculated animal groups (Fig. 4B). Our results indicate that the biofilm lifestyle reduced the ability of Salmonella to translocate to the spleen.
Host response induced by S. Enteritidis biofilm and planktonic bacteria.
Mice were infected intragastrically with 103 or 104 CFU of biofilm or planktonic S. Enteritidis. Studies were performed at day 5 after inoculation. (A) Bacterial burden in spleen and (B) spleen weight were analyzed. Results log CFU/g spleen and spleen weight are expressed as median, ns: non-significant; *p<0.05; **p<0.01; ***p<0.001. Mann–Whitney U test. (C) Splenic cytokine profile induced by S. Enteritidis. Cytokine expression was analyzed by qPCR. The relative mRNA amount was related to the mRNA levels of the control group defined as 1. Results are expressed as mean±SD. *p<0.05. ANOVA with Bonferroni correction. (D) Serum TNFα and IL-6 were measured by ELISA. Data are shown as mean±SD. *p<0.05. ANOVA with Bonferroni correction. (E and F) Intestinal histological changes. (E) Light microscopy of intestinal histology. H&E stain; 40× augmentation. Arrows indicate infiltration of inflammatory cells. (F) Intestinal inflammation scores. Lines indicate median values. **p<0.01. Mann–Whitney test. Five animals per group were analyzed. Representative data from three independent experiments.
Inflammatory response was investigated in systemic compartments: spleen and serum at day 5 post-infection. We found that spleens infected with planktonic Salmonella exhibited a significant increase in the expression of IL-17, IL-1β, IL-6, TNF-α and COX-2 compared with uninfected or Salmonella biofilm-infected organs (p<0.05). As shown in Figure 4C, 5 days post-inoculation with 104 CFU of planktonic bacteria, mice exhibited a 3-fold increase in the spleen expression of IL-17 and at least 1.5-fold increase in the expression of IL-1β, IL-6, TNF-α, and COX-2 compared with the biofilm infected group. Similar results but in a lower magnitude were observed when a 103 CFU dose per mouse was used. Likewise, significant differences were observed for TNF-α and IL-6 in serum between biofilm and planktonic inoculated mice (p<0.05, Fig. 4D). All animals inoculated with 103 CFU of Salmonella biofilm had similar levels of serum cytokines than the control group. As shown above, comparable spleen colonization (p>0.05) was detected when mice were inoculated with 104 CFU of Salmonella biofilm or 103 CFU of planktonic cells (median log CFU/g spleen: 3.54 and 3.65, respectively) (Fig. 4A). However, the biofilm inoculum was unable to generate an induction of inflammatory molecule expression as strong as that reached by its planktonic counterpart, even when the spleen bacterial burden was similar (Figs. 4A and C). These results suggest that the inflammatory host response to Salmonella varies depending on the bacterial lifestyle, being Salmonella biofilm less able to induce an inflammatory response than planktonic cells.
The host-response was also investigated in the murine gut. Results showed that animals infected with biofilm bacteria presented significantly reduced signs of intestinal inflammation compared with the planktonic group (p<0.01) (Figs. 4E and F). As expected, after ingestion of 103 CFU, planktonic Salmonella induced extended mucosal irritation. The intestinal epithelium showed a reduction in height; ileal villi were fused together and thickened, increasing in width because of the infiltration of inflammatory cells (score=between 2 and 3, Figs. 4E and F). In line with these results, 104 CFU of planktonic bacteria induced even a more pronounced pathology, with scores of 3 and 4. On the contrary, gut alterations induced by Salmonella biofilm were less injurious: the mice intestines infected with 104 CFU were characterized by focal mucosal irritation with poor loss of epithelial integrity, whereas with 103 CFU they showed edema with epithelial preservation. In summary, for the biofilm inoculum, the pathological scores ranged mainly between 1 or 2 independently of the inoculum of 103 and 104 CFU (Figs. 4E and F).
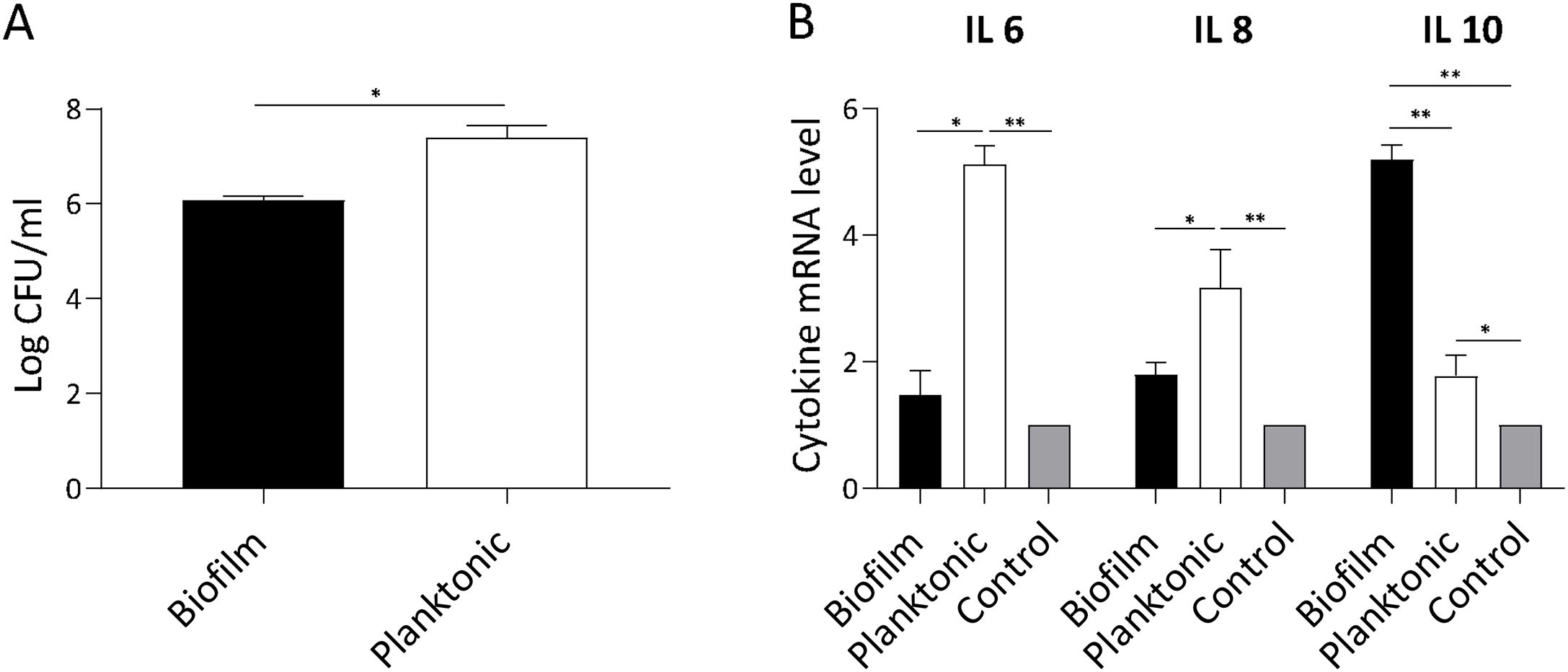
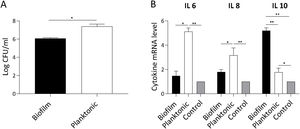
S. Enteritidis biofilm invasiveness and inflammatory response in non-phagocytic cellsWe assumed that attenuation of Salmonella biofilm might reflect an impaired bacterial interaction with host cells. Thus, we investigated whether biofilm and planktonic Salmonella differed in their invasion capacity of non-phagocytic cells. As shown in Figure 5A, Salmonella invasion capacity varied depending on the bacterial state. Using a similar MOI for both lifestyles, at 90min post-infection the number of intracellular bacteria recovered from cells infected with Salmonella biofilm was significantly lower (p<0.05) than that isolated from cells infected with planktonic bacteria (mean log CFU/ml: 6±SD vs. 7.4±SD, respectively). These data show that the Salmonella biofilm lifestyle is partially impaired for invading non-phagocytic cells. The inflammatory response of infected cells was also studied. As depicted in Fig. 5B, IL-6 expression induced by Salmonella biofilm was 3 times lower than that triggered by planktonic bacteria (p<0.05). In the case of IL-8, the level of expression induced by the biofilm was less than half the amount induced by the planktonic lifestyle (p<0.05). Moreover, no significant differences were observed in both cytokine expression between cells infected with the Salmonella biofilm and non-infected cells. Interestingly, Salmonella biofilm induced higher levels of anti-inflammatory cytokine IL-10 compared to planktonic cells and control (p<0.01, Fig. 5B). Taken together, these results indicate that the Salmonella biofilm lifestyle is less invasive and induces a reduced inflammatory response compared with its planktonic counterpart.
S. Enteritidis biofilm interaction with non- phagocytic cells. HEp-2 cells were infected with S. Enteritidis planktonic or biofilm lifestyle. Cells were processed 90min post-infection. Data shown are the mean±SD. Three independent experiments were performed. (A) Cell invasion capacity. Intracellular persisting bacteria were estimated by plating samples on TSA agar plates. (B) Cytokine expression. IL-6, IL-8 and IL-10 expression was determined by qPCR. The relative mRNA amount was related to the mRNA levels of the uninfected control group defined as 1. *p<0.05; **p<0.01. ANOVA with Bonferroni correction.
Salmonella adapts itself in extreme host and non-host environments both at the genetic and phenotypic level leading to their better survival and propagation18,24. In this regard, Salmonella strains might lose some virulence factors by in vitro passages in standard laboratory media. Over the past years, we have demonstrated that S. Enteritidis 5694 strain is highly virulent in the murine model of infection11,14,20. We observed by PCR and qPCR that S. Enteritidis 5694 expresses virulence genes such as: Salmonella Pathogenicity Island 1 (SPI-1) genes (invA, invG and sopA), sopE2 gene, a SPI-5 gene (sopB), a dual effector gene (sopD)20,21 (data not shown) and biofilm genes (this study). On this basis, S. Enteritidis 5694 strain was used to investigate the differential host response induced by S. Enteritidis biofilm and planktonic lifestyle.
S. Enteritidis is able to successfully survive in diverse milieus partly due to its ability to form biofilms. This ability has been involved in the non-typhoidal Salmonella persistence in the environment, and in medical, veterinary and industrial settings7,19. However, its function in the host-pathogen interactions has not been widely explored. It has been proposed that planktonic cells are adapted for virulence whereas biofilm cells are fit for persistence16. The link between virulence and persistence would be a strategy to improve the transmission probabilities each time that Salmonella leaves an infected host. This situation implies that the host immune system could react in a different way to these specialized cell types17. In fact, the results shown here reveal differences in the outcome of the interaction between S. Enteritidis and the host, depending on the bacterial lifestyle: planktonic or biofilm. In both murine and cellular models of infection, Salmonella biofilm was found to be less virulent than its planktonic counterpart.
After ingestion, Salmonella crosses the intestinal barrier in order to invade and colonize the intestinal tissues and other organs. Epithelial cells play an essential role in the result of infection by influencing the host inflammatory response26,27. We showed that a low dose of planktonic bacteria induces extended intestinal mucosal irritation with infiltration of inflammatory cells. Subsequently and in a successful manner, the pathogen induces splenic infection with 25% mortality. On the contrary, the limited effect of the Salmonella biofilm lifestyle inoculum observed on the intestinal epithelium and clinical features can be related to its drastically diminished colonization and spleen translocation capability. It is important to note that, even when the spleen bacterial burden was similar, the biofilm inoculum induces lower inflammatory response in serum and spleen than its planktonic counterpart. Other important aspects related to intestinal permeability are curli fibrils and cellulose, the components of Salmonella biofilms. Oppong et al.23 demonstrated that curli fibrils in S. Typhimurium play a beneficial host role regulating the intestinal barrier in addition to controlling the translocation of bacteria during infection. They suggest that bacterial amyloids reduce intestinal inflammation via the TLR-2/IL-10 axis. Ahmad et al.2 showed that a diminished cellulose production in S. Typhimurium reduces biofilm formation and enhances virulence. We determined that the Salmonella biofilm inoculum expresses higher levels of curli and cellulose genes with respect to the planktonic lifestyle. Based on our results and published data1,23, we hypothesize that Salmonella biofilm does not affect the gut barrier function as strongly as planktonic bacteria do, thus resulting in low bacterial translocation to the spleen and low mortality. Upon contact with the intestinal epithelial cells, Salmonella translocates bacterial effector proteins into the host cell cytosol using a Type Three Secretion System (T3SS) encoded by SPI-1, (T3SS-1). Once in the epithelial cell cytosol, Salmonella alters host cell signaling pathways that promote changes in the cytoskeleton, with consequent bacterial internalization and changes in inflammatory gene expression9. Our results indicate that murine infection with Salmonella biofilm cells is less virulent than its planktonic counterpart. We showed that Salmonella biofilm is less suitable to invade host cells. In this way, it has been previously demonstrated that T3SS-1 expression is increased in Salmonella planktonic cells, stimulating bacterial invasion, dissemination, and host inflammation. On the contrary, SPI-1 genes expression levels were reduced in biofilm cells17. MacKenzie et al.16 described that from the point of view of transmission, biofilm formation is beneficial for ensuring pathogen survival in the milieu. However, from the perspective of an infection, biofilm formation may be considered an anti-virulence feature. In summary, our results show that the outcome of Salmonella–host interaction varies depending on the bacterial lifestyle, where Salmonella biofilm is less invasive, less virulent and generates lower inflammatory response than its planktonic counterpart. In view of the fact that a predominant number of human infections are zoonotic in nature, our study contributes to a better understanding of the dynamic and complex interaction between Salmonella and the host. Results obtained from this line of work might allow to design new strategies to reduce the bacterial burden of livestock and human infections caused by this pathogen in the future, thus controlling economic losses.
Conflict of interestAuthors declare no conflict of interest.
This work was supported in part by grants from the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET)PUE 0085-2016, CONICET PIP 2015-2017 GI 11220150100383C and Universidad de Buenos Aires UBACyT 2018 N° 20020170100401BA.