The aim of this study was to compare the performance of two MALDI-TOF MS systems in the identification of clinically relevant strict anaerobic bacteria. The 16S rRNA gene sequencing was the gold standard method when discrepancies or inconsistencies were observed between platforms. A total of 333 isolates were recovered from clinical samples of different centers in Buenos Aires City between 2016 and 2021. The isolates were identified in duplicate using two MALDI-TOF MS systems, BD Bruker Biotyper (Bruker Daltonics, Bremen, Germany) and Vitek MS (bioMèrieux, Marcy-l’Etoile, France). Using the Vitek MS system, the identification of anaerobic isolates yielded the following percentages: 65.5% (n: 218) at the species or species–complex level, 71.2% (n: 237) at the genus level, 29.4% (n: 98) with no identification and 5.1% (n: 17) with misidentification. Using the Bruker Biotyper system, the identification rates were as follows: 85.3% (n: 284) at the species or species–complex level, 89.7% (n: 299) at the genus level, 14.1% (n: 47) with no identification and 0.6% (n: 2) with misidentification. Differences in the performance of both methods were statistically significant (p-values <0.0001). In conclusion, MALDI-TOF MS systems speed up microbial identification and are particularly effective for slow-growing microorganisms, such as anaerobic bacteria, which are difficult to identify by traditional methods. In this study, the Bruker system showed greater accuracy than the Vitek system. In order to be truly effective, it is essential to update the databases of both systems by increasing the number of each main spectrum profile within the platforms.
El objetivo de este estudio fue comparar el desempeño de dos sistemas MALDI-TOF MS en la identificación de bacterias anaerobias estrictas de interés clínico. La secuenciación del gen 16S ARNr fue el método de referencia utilizado cuando se observaron discrepancias o inconsistencias entre plataformas. Se recuperaron 333 aislados de muestras clínicas de diferentes centros de la Ciudad Autónoma de Buenos Aires entre 2016 y 2021. Los aislados se identificaron por duplicado mediante dos sistemas MALDI-TOF MS: el BD Bruker Biotyper (Bruker Daltonics, Bremen, Alemania) y el Vitek MS (bioMèrieux, Marcy-l’Etoile, Francia). A través del sistema Vitek MS, los mismos fueron identificados a nivel de especie o complejo de especies en un 65,5% (n: 218) y de género en un 71,2% (n: 237), mientras que no se identificaron en un 29,4% (n: 98) y fue incorrecta en el 5,1% (n: 17). Mediante el sistema Bruker Biotyper, dichos valores fueron del 85,3% (n: 284), del 89,7% (n: 299), del 14,1% (n: 47) y del 0,6% (n: 2), respectivamente. La diferencia entre ambos métodos fue estadísticamente significativa (p<0,0001). En conclusión, los sistemas MALDI-TOF MS aceleran la identificación microbiana. Son especialmente útiles para los microorganismos de crecimiento lento, como las bacterias anaerobias, que son difíciles de identificar con los métodos tradicionales. El sistema Bruker demostró ser más preciso que el Vitek MS. Para que estos métodos sean realmente efectivos es fundamental actualizar las bases de datos de ambos sistemas e incrementar el número de espectros de referencia dentro de las plataformas.
Matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) is a fast and accurate tool for the routine identification of microorganisms, being particularly effective for anaerobic bacteria, since their classical phenotypic identification requires long-term cultivation (at least 48h) and a substantial quantity of inoculum4,5,36,57,58. On the other hand, identification using MALDI-TOF MS requires low inoculum and takes only a few minutes. In this sense, this technology brought a new light to anaerobic microbiology and has been an important turning point4,5,17,36,57,58.
Numerous studies have described the efficacy of identifying anaerobic bacteria by MALDI-TOF MS, but most of them performed the identification using only one instrument (VITEK MS or Bruker Biotyper)2,38,55,82, and took into account the most common microorganisms that are routinely identified in clinical laboratories. Several authors concluded that there is a need to optimize and constantly update the existing MALDI-TOF MS databases8,26,75.
In Argentina, the commercially available MALDI-TOF MS systems used for the identification of microorganisms isolated from clinical specimens are MALDI-TOF Biotyper (Bruker Daltonics, Bremen, Germany), and VITEK MS (bioMèrieux, Marcy l’Etoile, France). Scarce literature compared the efficiency of both platforms in the identification of anaerobic bacteria33,34,39,52.
The aim of this study was to compare the performances of two MALDI-TOF MS systems in the identification of clinical strict anaerobic isolates, using the sequence analysis of the 16S ribosomal RNA (16S rRNA) gene as the gold standard method when discrepancies or inconsistencies were observed between these two mass spectrometry methods.
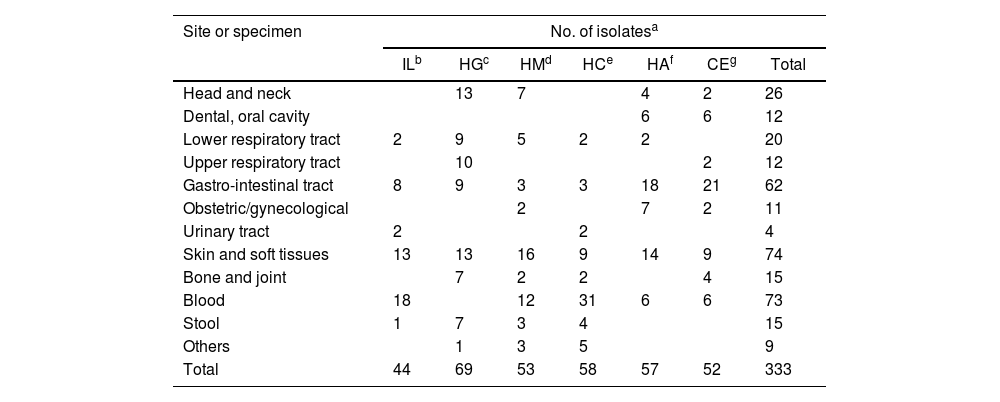
Materials and methodsBacterial isolatesA total of 333 isolates were recovered from clinical samples collected from six different centers in Buenos Aires City between 2016 and 2021 (Table 1). Gram staining of the recovered anaerobic isolates was performed, and an oxygen tolerance test was conducted to ensure culture purity. Identification was carried out by phenotypic methods as previously described53. Using phenotypic methods in several isolates we achieved genus level identification every time45. Isolates were stored at −70°C in trypticase soy broth with 20% glycerol. Frozen isolates were then sub-cultured on Brucella blood agar and incubated for 48h under an anaerobic atmosphere before MALDI-TOF MS identification.
Distribution of sources and isolates from six centers.
| Site or specimen | No. of isolatesa | ||||||
|---|---|---|---|---|---|---|---|
| ILb | HGc | HMd | HCe | HAf | CEg | Total | |
| Head and neck | 13 | 7 | 4 | 2 | 26 | ||
| Dental, oral cavity | 6 | 6 | 12 | ||||
| Lower respiratory tract | 2 | 9 | 5 | 2 | 2 | 20 | |
| Upper respiratory tract | 10 | 2 | 12 | ||||
| Gastro-intestinal tract | 8 | 9 | 3 | 3 | 18 | 21 | 62 |
| Obstetric/gynecological | 2 | 7 | 2 | 11 | |||
| Urinary tract | 2 | 2 | 4 | ||||
| Skin and soft tissues | 13 | 13 | 16 | 9 | 14 | 9 | 74 |
| Bone and joint | 7 | 2 | 2 | 4 | 15 | ||
| Blood | 18 | 12 | 31 | 6 | 6 | 73 | |
| Stool | 1 | 7 | 3 | 4 | 15 | ||
| Others | 1 | 3 | 5 | 9 | |||
| Total | 44 | 69 | 53 | 58 | 57 | 52 | 333 |
The isolates were identified in duplicate using the two MALDI-TOF MS systems. Bacterial isolates were identified by the direct colony on-plate extraction method with the Bruker Biotyper system, using the MALDI Biotyper software 3.1 (library version 10.0 containing 9607 main spectrum profile [MSP] entries), as previously described69.
The cut-off score used for identification using Bruker Biotyper was ≥1.5 for the genus level, and ≥1.7 for the species level. Otherwise, the identification was considered unreliable. A minimum difference of 10% between the top score and the next closest score was required for the bacterial isolate to be considered a different species, based on the interpretative criteria of the National Network for Microbiological Identification by Mass Spectrometry (RENAEM) (http://www.anlis.gov.ar/renaem/)32,56.
Identification was carried out using the Vitek MS system with the platform v3.2 knowledge base for clinical use. All procedures followed the manufacturer's instructions. Values between 60.0 and 99.9% indicated reliable species discrimination.
To compare system performance, MALDI-TOF MS results were classified into four categories: 1 – correct identification of species or species–complex; 2 – correct identification of genus; 3 – no identification, and 4 – misidentification. The species whose names were updated in recent years and assigned by MALDI-TOF MS with their previous name were considered correct identification. Rare species were defined as such when fewer than 10 articles related to the subject were retrieved from the PubMed database (https://pubmed.ncbi.nlm.nih.gov/)36.
Genetic identificationThe sequence analysis of 16S rRNA gene was used as a reference method when: (1) isolates could not be identified by any MALDI-TOF MS, (2) isolate identifications showed discrepancies between both systems either because they were different or one of the platforms yielded a “no identification” result and (3) identification was unreliable, that is, when the percentage/score of identification fell below the cut-off established by any system34.
Genomic DNA was extracted using a commercial preparation kit (ADN PuriPrep-B, Inbio Highway), and the 16S rRNA gene was amplified using the specific primers 63f (5′-CAGGCCTAACACATGCAAGTC-3′) and 1387r (5′-GGGCGGWGTGTACAAGGC-3′). Amplicons were sequenced at external facilities (Macrogen, South Korea). Sequences obtained were analyzed using the Blastn online tool at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST) and the VECTOR NTI 11.5 program.
To achieve the identification at species level the sequences should exhibit an identity of >99% with full gene coverage (>80%) with 16S rRNA gene of reference sequences based on guideline MM18-A of the Clinical and Laboratory Standards Institute (CLSI)12. However, for Bacteroidesthetaiotaomicron/Bacteroides faecis, Bacteroides ovatus/Bacteroides xylanisolvens, Fusobacterium nucleatum/Fusobacterium naviforme, Porphyromonas asaccharolytica/Porphyromonas uenonis and Peptoniphilus harei/Peptoniphilus indolicus, the identification was considered correct as a complex, since they could not be differentiated by 16S rRNA sequencing14.
Statistical analysisConfidence interval, proportions and differences between platforms were calculated by the proportion methods using Statistix 10.0. Statistical significance was assigned to p-values <0.05.
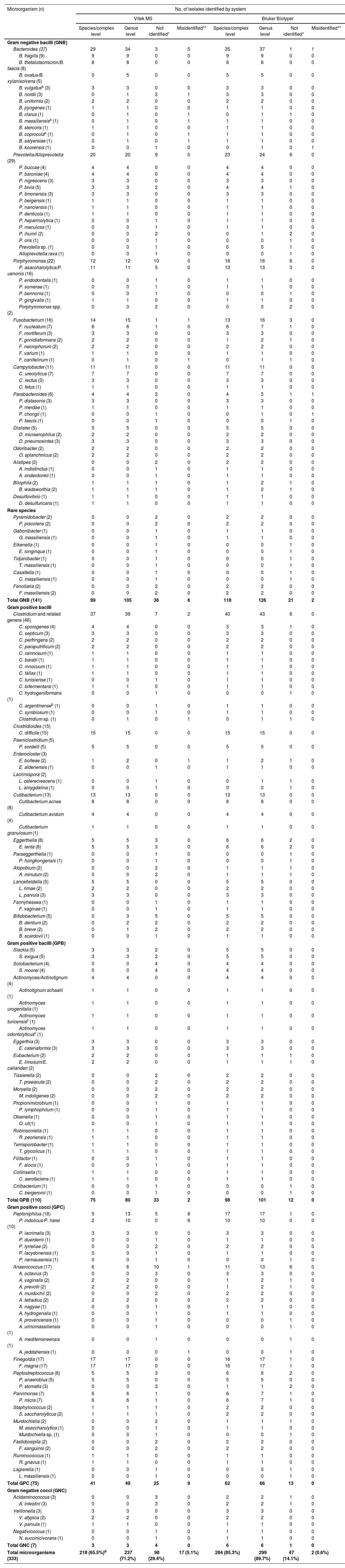
Results and discussionTable 2 summarizes the identification of the 333 clinical anaerobic isolates, with 141 isolates corresponding to gram negative bacilli (GNB),110 isolates to gram positive bacilli (GPB), 75 isolates to gram positive cocci (GPC), and 7 isolates to gram negative cocci (GNC) distributed among 58 different genera and 139 species, many of which are recognized as causes of human infections.
Identification of 333 anaerobic bacteria by the Vitek MS and Bruker Biotyper systems.
| Microorganism (n) | No. of isolates identified by system | |||||||
|---|---|---|---|---|---|---|---|---|
| Vitek MS | Bruker Biotyper | |||||||
| Species/complex level | Genus level | Not identified* | Misidentified** | Species/complex level | Genus level | Not identified* | Misidentified** | |
| Gram negative bacilli (GNB) | ||||||||
| Bacteroides (37) | 29 | 34 | 3 | 5 | 35 | 37 | 1 | 1 |
| B. fragilis (9) | 9 | 9 | 0 | 0 | 9 | 9 | 0 | 0 |
| B. thetaiotaomicron/B. faecis (8) | 8 | 8 | 0 | 0 | 8 | 8 | 0 | 0 |
| B. ovatus/B. xylanisolvens (5) | 5 | 5 | 0 | 0 | 5 | 5 | 0 | 0 |
| B. vulgatusa (3) | 3 | 3 | 0 | 0 | 3 | 3 | 0 | 0 |
| B. nordii (3) | 0 | 1 | 2 | 1 | 3 | 3 | 0 | 0 |
| B. uniformis (2) | 2 | 2 | 0 | 0 | 2 | 2 | 0 | 0 |
| B. pyogenes (1) | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 |
| B. clarus (1) | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 0 |
| B. massiliensisa (1) | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 0 |
| B. stercoris (1) | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 |
| B. coprocolaa (1) | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 0 |
| B. salyersiae (1) | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 0 |
| B. koorensis (1) | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 |
| Prevotella/Alloprevotella (29) | 20 | 20 | 9 | 0 | 23 | 24 | 6 | 0 |
| P. buccae (4) | 4 | 4 | 0 | 0 | 4 | 4 | 0 | 0 |
| P. baroniae (4) | 4 | 4 | 0 | 0 | 4 | 4 | 0 | 0 |
| P. nigrescens (3) | 3 | 3 | 0 | 0 | 3 | 3 | 0 | 0 |
| P. bivia (5) | 3 | 3 | 2 | 0 | 4 | 4 | 1 | 0 |
| P. timonensis (3) | 3 | 3 | 0 | 0 | 3 | 3 | 0 | 0 |
| P. bergensis (1) | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 |
| P. nanciensis (1) | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 |
| P. denticola (1) | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 |
| P. heparinolytica (1) | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 |
| P. maculosa (1) | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 |
| P. ihumii (2) | 0 | 0 | 2 | 0 | 0 | 1 | 2 | 0 |
| P. oris (1) | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| Prevotella sp. (1) | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| Alloprevotella rava (1) | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| Porphyromonas (22) | 12 | 12 | 10 | 0 | 16 | 16 | 6 | 0 |
| P. asaccharolytica/P. uenonis (16) | 11 | 11 | 5 | 0 | 13 | 13 | 3 | 0 |
| P. endodontalis (1) | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 |
| P. somerae (1) | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 |
| P. bennonis (1) | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| P. gingivalis (1) | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 |
| Porphyromonas spp. (2) | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 |
| Fusobacterium (16) | 14 | 15 | 1 | 1 | 13 | 16 | 3 | 0 |
| F. nucleatum (7) | 6 | 6 | 1 | 0 | 6 | 7 | 1 | 0 |
| F. mortiferum (3) | 3 | 3 | 0 | 0 | 3 | 3 | 0 | 0 |
| F. gonidiaformans (2) | 2 | 2 | 0 | 0 | 1 | 2 | 1 | 0 |
| F. necrophorum (2) | 2 | 2 | 0 | 0 | 2 | 2 | 0 | 0 |
| F. varium (1) | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 |
| F. canifelinum (1) | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 0 |
| Campylobacter (11) | 11 | 11 | 0 | 0 | 11 | 11 | 0 | 0 |
| C. ureolyticus (7) | 7 | 7 | 0 | 0 | 7 | 7 | 0 | 0 |
| C. rectus (3) | 3 | 3 | 0 | 0 | 3 | 3 | 0 | 0 |
| C. fetus (1) | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 |
| Parabacteroides (6) | 4 | 4 | 2 | 0 | 4 | 5 | 1 | 1 |
| P. distasonis (3) | 3 | 3 | 0 | 0 | 3 | 3 | 0 | 0 |
| P. merdae (1) | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 |
| P. chongii (1) | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 |
| P. faecis (1) | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| Dialister (5) | 5 | 5 | 0 | 0 | 5 | 5 | 0 | 0 |
| D. micraerophilus (2) | 2 | 2 | 0 | 0 | 2 | 2 | 0 | 0 |
| D. pneumosintes (3) | 3 | 3 | 0 | 0 | 3 | 3 | 0 | 0 |
| Odoribacter (2) | 2 | 2 | 0 | 0 | 2 | 2 | 0 | 0 |
| O. splanchnicus (2) | 2 | 2 | 0 | 0 | 2 | 2 | 0 | 0 |
| Alistipes (2) | 0 | 0 | 2 | 0 | 2 | 2 | 0 | 0 |
| A. indistinctus (1) | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 |
| A. onderdonkii (1) | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 |
| Bilophila (2) | 1 | 1 | 1 | 0 | 1 | 2 | 1 | 0 |
| B. wadsworthia (2) | 1 | 1 | 1 | 0 | 1 | 2 | 1 | 0 |
| Desulfovibrio (1) | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 |
| D. desulfuricans (1) | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 |
| Rare species | ||||||||
| Pyramidobacter (2) | 0 | 0 | 2 | 0 | 2 | 2 | 0 | 0 |
| P. piscolens (2) | 0 | 0 | 2 | 0 | 2 | 2 | 0 | 0 |
| Gabonibacter (1) | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 |
| G. massiliensis (1) | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 |
| Eikenella (1) | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| E. longinqua (1) | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| Tidjanibacter (1) | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| T. massiliensis (1) | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| Casaltella (1) | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| C. massiliensis (1) | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| Fenollaria (2) | 0 | 0 | 2 | 0 | 2 | 2 | 0 | 0 |
| F. massiliensis (2) | 0 | 0 | 2 | 0 | 2 | 2 | 0 | 0 |
| Total GNB (141) | 99 | 105 | 36 | 6 | 118 | 126 | 21 | 2 |
| Gram positive bacilli | ||||||||
| Clostridium and related genera (46) | 37 | 39 | 7 | 2 | 40 | 43 | 6 | 0 |
| C. sporogenes (4) | 4 | 4 | 0 | 0 | 3 | 3 | 1 | 0 |
| C. septicum (3) | 3 | 3 | 0 | 0 | 3 | 3 | 0 | 0 |
| C. perfringens (2) | 2 | 2 | 0 | 0 | 2 | 2 | 0 | 0 |
| C. paraputrificum (2) | 2 | 2 | 0 | 0 | 2 | 2 | 0 | 0 |
| C. ramnosum (1) | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 |
| C. baratii (1) | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 |
| C. innocuum (1) | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 |
| C. fallax (1) | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 |
| C. tunisiense (1) | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 |
| C. bifermentans (1) | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 |
| C. hydrogeniformans (1) | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| C. argentinenseb (1) | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 |
| C. symbiosum (1) | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 |
| Clostridium sp. (1) | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 0 |
| Clostridioides (15) | ||||||||
| C. difficile (15) | 15 | 15 | 0 | 0 | 15 | 15 | 0 | 0 |
| Paeniclostridium (5) | ||||||||
| P. sordelli (5) | 5 | 5 | 0 | 0 | 5 | 5 | 0 | 0 |
| Enterocloster (3) | ||||||||
| E. bolteae (2) | 1 | 2 | 0 | 1 | 1 | 2 | 1 | 0 |
| E. aldenensis (1) | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 |
| Lacrimispora (2) | ||||||||
| L. celerecrescens (1) | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 |
| L. amygdalina (1) | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| Cutibacterium (13) | 13 | 13 | 0 | 0 | 13 | 13 | 0 | 0 |
| Cutibacterium acnes (8) | 8 | 8 | 0 | 0 | 8 | 8 | 0 | 0 |
| Cutibacterium avidum (4) | 4 | 4 | 0 | 0 | 4 | 4 | 0 | 0 |
| Cutibacterium granulosum (1) | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 |
| Eggerthella (8) | 5 | 5 | 3 | 0 | 6 | 6 | 2 | 0 |
| E. lenta (8) | 5 | 5 | 3 | 0 | 6 | 6 | 2 | 0 |
| Paraeggerthella (1) | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| P. hongkongensis (1) | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| Atopobium (2) | 0 | 0 | 2 | 0 | 1 | 1 | 1 | 0 |
| A. minutum (2) | 0 | 0 | 2 | 0 | 1 | 1 | 1 | 0 |
| Lancefieldella (5) | 5 | 5 | 0 | 0 | 5 | 5 | 0 | 0 |
| L. rimae (2) | 2 | 2 | 0 | 0 | 2 | 2 | 0 | 0 |
| L. parvula (3) | 3 | 3 | 0 | 0 | 3 | 3 | 0 | 0 |
| Fannyhessea (1) | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 |
| F. vaginae (1) | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 |
| Bifidobacterium (5) | 0 | 3 | 5 | 0 | 5 | 5 | 0 | 0 |
| B. dentium (2) | 0 | 2 | 2 | 0 | 2 | 2 | 0 | 0 |
| B. breve (2) | 0 | 1 | 2 | 0 | 2 | 2 | 0 | 0 |
| B. scardovii (1) | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 |
| Gram positive bacilli (GPB) | ||||||||
| Slackia (5) | 3 | 3 | 2 | 0 | 5 | 5 | 0 | 0 |
| S. exigua (5) | 3 | 3 | 2 | 0 | 5 | 5 | 0 | 0 |
| Solobacterium (4) | 0 | 0 | 4 | 0 | 4 | 4 | 0 | 0 |
| S. moorei (4) | 0 | 0 | 4 | 0 | 4 | 4 | 0 | 0 |
| Actinomyces/Actinotignum (4) | 4 | 4 | 0 | 0 | 4 | 4 | 0 | 0 |
| Actinotignum schaalii (1) | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 |
| Actinomyces urogenitalis (1) | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 |
| Actinomyces turicensisc (1) | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 |
| Actinomyces odontolyticusc (1) | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 |
| Eggerthia (3) | 3 | 3 | 0 | 0 | 3 | 3 | 0 | 0 |
| E. catenaformis (3) | 3 | 3 | 0 | 0 | 3 | 3 | 0 | 0 |
| Eubacterium (2) | 2 | 2 | 0 | 0 | 1 | 1 | 1 | 0 |
| E. limosum/E. callanderi (2) | 2 | 2 | 0 | 0 | 1 | 1 | 1 | 0 |
| Tissierella (2) | 0 | 0 | 2 | 0 | 2 | 2 | 0 | 0 |
| T. praeacuta (2) | 0 | 0 | 2 | 0 | 2 | 2 | 0 | 0 |
| Moryella (2) | 0 | 0 | 2 | 0 | 2 | 2 | 0 | 0 |
| M. indoligenes (2) | 0 | 0 | 2 | 0 | 2 | 2 | 0 | 0 |
| Propionimicrobium (1) | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 |
| P. lymphophilum (1) | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 |
| Olsenella (1) | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 |
| O. uli(1) | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 |
| Robinsoniella (1) | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 |
| R. peoriensis (1) | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 |
| Terrisporobacter (1) | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 |
| T. glycolicus (1) | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 |
| Filifactor (1) | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 |
| F. alocis (1) | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 |
| Collinsella (1) | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 |
| C. aerofaciens (1) | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 |
| Criibacterium (1) | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| C. bergeronii (1) | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| Total GPB (110) | 75 | 80 | 33 | 2 | 98 | 101 | 12 | 0 |
| Gram positive cocci (GPC) | ||||||||
| Peptoniphilus (18) | 5 | 13 | 5 | 8 | 17 | 17 | 1 | 0 |
| P. indolicus/P. harei (10) | 2 | 10 | 0 | 8 | 10 | 10 | 0 | 0 |
| P. lacrimalis (3) | 3 | 3 | 0 | 0 | 3 | 3 | 0 | 0 |
| P. duerdenii (1) | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 |
| P. tyrreliae (2) | 0 | 0 | 2 | 0 | 2 | 2 | 0 | 0 |
| P. lacydonensis (1) | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 |
| P. nemausensis (1) | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| Anaerococcus (17) | 6 | 6 | 10 | 1 | 11 | 13 | 6 | 0 |
| A. octavius (3) | 0 | 0 | 3 | 0 | 3 | 3 | 0 | 0 |
| A. vaginalis (2) | 2 | 2 | 0 | 0 | 1 | 2 | 1 | 0 |
| A. prevotii (2) | 2 | 2 | 0 | 0 | 1 | 2 | 1 | 0 |
| A. murdochii (2) | 0 | 0 | 2 | 0 | 2 | 2 | 0 | 0 |
| A. tetradius (2) | 2 | 2 | 0 | 0 | 2 | 2 | 0 | 0 |
| A. nagyae (1) | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 |
| A. hydrogenalis (1) | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 |
| A. provencensis (1) | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| A. urinomassiliensis (1) | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| A. mediterraneensis (1) | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| A. jeddahensis (1) | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 |
| Finegoldia (17) | 17 | 17 | 0 | 0 | 16 | 17 | 1 | 0 |
| F. magna (17) | 17 | 17 | 0 | 0 | 16 | 17 | 1 | 0 |
| Peptostreptococcus (8) | 5 | 5 | 3 | 0 | 6 | 6 | 2 | 0 |
| P. anaerobius (5) | 5 | 5 | 0 | 0 | 5 | 5 | 0 | 0 |
| P. stomatis (3) | 0 | 0 | 3 | 0 | 1 | 1 | 2 | 0 |
| Parvimonas (7) | 6 | 6 | 1 | 0 | 6 | 7 | 1 | 0 |
| P. micra (7) | 6 | 6 | 1 | 0 | 6 | 7 | 1 | 0 |
| Staphylococcus (2) | 1 | 1 | 1 | 0 | 2 | 2 | 0 | 0 |
| S. saccharolyticus (2) | 1 | 1 | 1 | 0 | 2 | 2 | 0 | 0 |
| Murdochiella (2) | 0 | 0 | 2 | 0 | 1 | 1 | 1 | 0 |
| M. asaccharolytica (1) | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 |
| Murdochiella sp. (1) | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| Fastidiosipila (2) | 0 | 0 | 2 | 0 | 2 | 2 | 0 | 0 |
| F. sanguinis (2) | 0 | 0 | 2 | 0 | 2 | 2 | 0 | 0 |
| Ruminococcus (1) | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 |
| R. gnavus (1) | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 |
| Lagierella (1) | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| L. massiliensis (1) | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| Total GPC (75) | 41 | 49 | 25 | 9 | 62 | 66 | 13 | 0 |
| Gram negative cocci (GNC) | ||||||||
| Acidaminococcus (3) | 0 | 0 | 3 | 0 | 2 | 2 | 1 | 0 |
| A. intestini (3) | 0 | 0 | 3 | 0 | 2 | 2 | 1 | 0 |
| Veillonella (3) | 3 | 3 | 0 | 0 | 3 | 3 | 0 | 0 |
| V. atypica (2) | 2 | 2 | 0 | 0 | 2 | 2 | 0 | 0 |
| V. parvula (1) | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 |
| Negativicoccus (1) | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 |
| N. succinicivorans (1) | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 |
| Total GNC (7) | 3 | 3 | 4 | 0 | 6 | 6 | 1 | 0 |
| Total microorganisms (333) | 218 (65.5%)d | 237 (71.2%) | 98 (29.4%) | 17 (5.1%) | 284 (85.3%) | 299 (89.7%) | 47 (14.1%) | 2 (0.6%) |
In this study, the identification at the species or species–complex level comprised 85.3% (n: 284/333) (CI95: 81.3–98.2) of the isolates using the Bruker System, but Vitek MS achieved less efficacy, given that only 65.5% (n: 218/333) (CI95: 60.2–70.7) of the isolates could be identified (p-value <0.0001) (Table 2). Previous studies reported that correct identification at the species level using the Bruker Biotyper platform ranged from 70.8 to 95.7%, while the Vitek MS platform reached 82.2–91.2% of the isolates2,26,38,45,57,65,75.
- -
Identification of gram negative bacilli (GNB) isolates
The performances in the identifications are shown in Tables 2 and 3. Of the GNB, 89.4% (126/141) and 74.5% (105/141) were identified at the genus level using the Bruker Biotyper system and the Vitek MS System, respectively (p-value: 0.002).
Overall picture of the identification of the species included in this work and their presence in databases.
| Microorganism | Spectra included in | Identified, misidentified, or not identified at the species level by | ||
|---|---|---|---|---|
| Vitek system | Bruker Biotyper system | Vitek system | Bruker Biotyper system | |
| Gram negative bacilli (GNB) | ||||
| Bacteroides | ||||
| B. fragilis | x | x | Identifiedb | Identified |
| B. thetaiotaomicron/B. faecis | x | x | Identified | Identified |
| B. ovatus/B. xylanisolvens | x | xa | Identified | Identified |
| B. vulgatus* | x | x | Identified | Identified |
| B. nordii | – | x | Not Identified/misidentifiedc | Identified |
| B. uniformis | x | x | Identified | Identified |
| B. pyogenes | x | x | Identified | Identified |
| B. clarus | – | x | Misidentifiedd | Identified |
| B. massiliensis* | – | x | Misidentified | Identified |
| B. stercoris | x | x | Identified | Identified |
| B. coprocola* | – | x | Misidentified | Identified |
| B. salyersiae | – | x | Misidentified | Identified |
| B. koorensis | – | – | Not identifiede | Misidentified |
| Prevotella/Alloprevotella | ||||
| P. buccae | x | x | Identified | Identified |
| P. baroniae | x | x | Identified | Identified |
| P. nigrescens | x | x | Identified | Identified |
| P. bivia | x | x | Identified/Not identifiedf | Identified/Not identified |
| P. timonensis | x | x | Identified | Identified |
| P. bergensis | x | x | Identified | Identified |
| P. nanciensis | x | x | Identified | Identified |
| P. denticola | x | x | Identified | Identified |
| P. heparinolytica | – | x | Not identified | Identified |
| P. maculosa | – | x | Not identified | Identified |
| P. ihumii | – | – | Not identified | Not identified |
| P. oris | x | x | Not identified | Not identified |
| Alloprevotella rava | – | x | Not identified | Not identified |
| Porphyromonas | ||||
| P. asaccharolytica/P. uenonis | x | x | Identified/Not identified | Identified/Not identified |
| P. endodontalis | – | x | Not identified | Identified |
| P. somerae | – | x | Not identified | Identified |
| P. bennonis | – | x | Not identified | Not identified |
| P. gingivalis | x | x | Identified | Identified |
| Fusobacterium | ||||
| F. nucleatum | x | x | Identified/Not identified | Identified/Not identified |
| F. mortiferum | x | x | Identified | Identified |
| F. gonidiaformans | x | x | Identified | Identified/Not identified |
| F. necrophorum | x | x | Identified | Identified |
| F. varium | x | x | Identified | Identified |
| F. canifelinum | – | x | Misidentified | Not identified |
| Campylobacter | ||||
| C. ureolyticus | x | x | Identified | Identified |
| C. rectus | x | x | Identified | Identified |
| C. fetus | x | x | Identified | Identified |
| Parabacteroides | ||||
| P. distasonis | x | x | Identified | Identified |
| P. merdae | x | x | Identified | Identified |
| P. chongii | – | – | Not identified | Misidentified |
| P. faecis | – | – | Not identified | Not identified |
| Dialister | ||||
| D. micraerophilus | x | x | Identified | Identified |
| D. pneumosintes | x | x | Identified | Identified |
| Odoribacter | ||||
| O. splanchnicus | x | x | Identified | Identified |
| Alistipes | ||||
| A. indistinctus | – | x | Not identified | Identified |
| A. onderdonkii | – | x | Not identified | Identified |
| Bilophila | ||||
| B. wadsworthia | x | x | Identified/Not identified | Identified/Not identified |
| Desulfovibrio | ||||
| D. desulfuricans | x | x | Identified | Identified |
| Rare species | ||||
| Pyramidobacter | ||||
| P. piscolens | – | x | Not identified | Identified |
| Gabonibacter | ||||
| G. massiliensis | – | x | Not identified | Identified |
| Eikenella | ||||
| E. longinqua | – | – | Not identified | Not identified |
| Tidjanibacter | ||||
| T. massiliensis | – | – | Not identified | Not identified |
| Casaltella | ||||
| C. massiliensis | – | – | Not identified | Not identified |
| Fenollaria | ||||
| F. massiliensis | – | x | Not identified | Identified |
| Gram positive bacilli (GPB) | ||||
| Clostridium | ||||
| C. sporogenes | x | x | Identified | Identified/Not identified |
| C. septicum | x | x | Identified | Identified |
| C. perfringens | x | x | Identified | Identified |
| C. paraputrificum | x | x | Identified | Identified |
| C. ramnosum | x | x | Identified | Identified |
| C. baratii | x | x | Identified | Identified |
| C. innocuum | x | x | Identified | Identified |
| C. fallax | x | x | Identified | Identified |
| C. tunisiense | – | x | Not identified | Identified |
| C. bifermentans | x | x | Identified | Identified |
| C. hydrogeniformans | – | – | Not identified | Not identified |
| C. argentinense** | – | x | Not identified | Identified |
| C. symbiosum | – | x | Not identified | Identified |
| Clostridioides | ||||
| C. difficile | x | x | Identified | Identified |
| Paeniclostridium | ||||
| P. sordelli | x | x | Identified | Identified |
| Enterocloster | ||||
| E. bolteae | x | x | Identified/Misidentifiedg | Identified/Not identified |
| E. aldenensis | – | x | Not identified | Identified |
| Lacrimispora | ||||
| L. celerecrescens | – | x | Not identified | Not identified |
| L. amygdalina | – | – | Not identified | Not identified |
| Cutibacterium | ||||
| Cutibacterium acnes | x | x | Identified | Identified |
| Cutibacterium avidum | x | x | Identified | Identified |
| Cutibacterium granulosum | x | x | Identified | Identified |
| Eggerthella | ||||
| E. lenta | x | x | Identified/Not identified | Identified/Not identified |
| Paraeggerthella | ||||
| P. hongkongensis | – | – | Not identified | Not identified |
| Atopobium | ||||
| A. minutum | – | x | Not identified | Identified/Not identified |
| Lancefieldella | ||||
| L. rimae | x | x | Identified | Identified |
| L. parvula | x | x | Identified | Identified |
| Fannyhessea | ||||
| F. vaginae | – | x | Not identified | Identified |
| Bifidobacterium | ||||
| B. dentium | x | x | Not identified | Identified |
| B. breve | x | x | Not identified | Identified |
| B. scardovii | x | x | Not identified | Identified |
| Slackia | ||||
| S. exigua | x | x | Identified/Not identified | Identified |
| Solobacterium | ||||
| S. moorei | – | x | Not identified | Identified |
| Actinomyces/Actinotignum | ||||
| Actinotignum schaalii | x | x | Identified | Identified |
| Actinomyces turicensis*** | x | x | Identified | Identified |
| Actinomyces odontolyticus*** | x | x | Identified | Identified |
| Actinomyces urogenitalis | x | x | Identified | Identified |
| Eggerthia | ||||
| E. catenaformis | x | x | Identified | Identified |
| Eubacterium | ||||
| E. limosum/E. callanderi | x | x | Identified | Identified/Not identified |
| Tissierella | ||||
| T. praeacuta | – | x | Not identified | Identified |
| Moryella | ||||
| M. indoligenes | – | x | Not identified | Identified |
| Olsenella | ||||
| O. uli | – | x | Not identified | Identified |
| Robinsoniella | ||||
| R. peoriensis | x | x | Identified | Identified |
| Terrisporobacter | ||||
| T. glycolicus | x | x | Identified | Identified |
| Filifactor | ||||
| F. alocis | – | x | Not identified | Identified |
| Collinsella | ||||
| C. aerofaciens | x | x | Identified | Identified |
| Criibacterium | ||||
| C. bergeronii | – | – | Not identified | Not identified |
| Gram positive cocci (GPC) | ||||
| Peptoniphilus | ||||
| P. indolicus/P. harei | x | x | Identified/Misidentified | Identified |
| P. lacrimalis | x | x | Identified | Identified |
| P. duerdenii | – | x | Not identified | Identified |
| P. tyrreliae | – | x | Not identified | Identified |
| P. lacydonensis | – | x | Not identified | Identified |
| P. nemausensis | – | – | Not identified | Not identified |
| Anaerococcus | ||||
| A. octavius | – | x | Not identified | Identified |
| A. vaginalis | x | x | Identified | Identified/Not identified |
| A. prevotii | x | x | Identified | Identified/Not identified |
| A. murdochii | – | x | Not identified | Identified |
| A. tetradius | x | x | Identified | Identified |
| A. nagyae | – | x | Not identified | Identified |
| A. hydrogenalis | – | x | Not identified | Identified |
| A. provencensis | – | – | Not identified | Not identified |
| A. urinomassiliensis | – | – | Not identified | Not identified |
| A. mediterraneensis | – | – | Not identified | Not identified |
| A. jeddahensis | – | – | Misidentified | Not identified |
| Finegoldia | ||||
| F. magna | x | x | Identified | Identified/Not identified |
| Peptostreptococcus | ||||
| P. anaerobius | x | x | Identified | Identified |
| P. stomatis | – | x | Not identified | Identified/Not identified |
| Parvimonas | ||||
| P. micra | x | x | Identified/Not identified | Identified/Not identified |
| Staphylococcus | ||||
| S. saccharolyticus | x | x | Identified/Not identified | Identified |
| Murdochiella | ||||
| M. asaccharolytica | – | x | Not identified | Identified |
| Fastidiosipila | ||||
| F. sanguinis | – | X | Not identified | Identified |
| Ruminococcus | ||||
| R. gnavus | x | X | Identified | Identified |
| Lagierella | ||||
| L. massiliensis | – | – | Not identified | Not identified |
| Gram negative cocci (GNC) | ||||
| Acidaminococcus | ||||
| A. intestini | – | X | Not identified | Identified/Not identified |
| Veillonella | ||||
| V. atypica | x | X | Identified | Identified |
| V. parvula | x | X | Identified | Identified |
| Negativicoccus | ||||
| N. succinicivorans | – | X | Not identified | Identified |
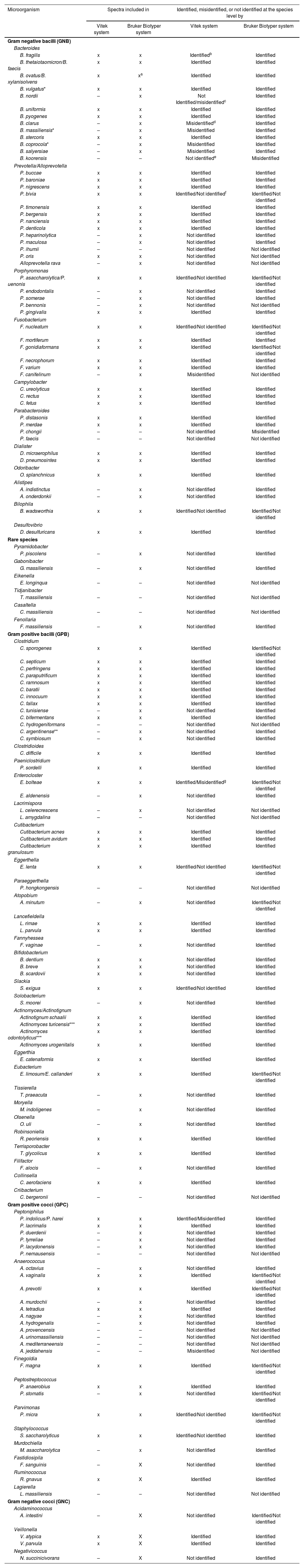
Notes: x, spectra included in system; –, spectra not included in system.
The GNB belonged to the following genera: Bacteroides, Porphyromonas, Prevotella, Alloprevotella, Parabacteroides, Alistipes, Odoribacter, Gabonibacter, Fusobacterium, Campylobacter, Dialister, Bilophila, Desulfovibrio, Pyramidobacter, Eikenella, Tidjanibacter and Casaltella, which were distributed into the eight different classes and orders and Fenollaria (S1).
At the species level, 83.7% (118/141) of GNB were identified by Bruker Biotyper, while Vitek MS identified 70.2% (99/141) (p-value: 0.011). The proportion of unidentified GNB was 14.9% (21/141) and 25.5% (36/141) by Bruker Biotyper and Vitek MS, respectively (p-value: 0.038). Both systems misidentified these bacteria in 1.4% by Bruker Biotyper, and in 4.3% by Vitek MS (p-value: 0.282). GNB not reliably identified mainly corresponded to the Porphyromonas genus followed by the Prevotella genus.
- ∘
Class Bacteroidia. Order Bacteroidales
Regarding Bacteroides spp., Vitek MS system identified 34/37 isolates at the genus level and showed difficulties in the discrimination of 8 isolates at the species level (Tables 2 and 3). For the following species: Bacteroides coprocola, Bacteroides salyersiae, Bacteroides nordii, and Bacteroides clarus, the identification failed since they are not included in the database.
Bruker Biotyper identified all Bacteroides isolates at the genus level, but one (L5) was not identified at the species level since the score difference between the two species was less than 10% (Table 4), and another isolate was misidentified at the species level.
Comparison between identifications provided by both MALDI-TOF MS systems and the reference method for the isolates with discrepancies, or with the same identification but low scores.
| Isolate/number | Vitek MS ID 99% | Bruker Biotyper ID (score) | 16S rRNA sequencing |
|---|---|---|---|
| Gram negative bacilli (GNB) | |||
| Bacteroides (n: 10) | |||
| L5 | B. stercoris | B. clarus (2.34), B. stercoris (2.23) | B. clarus |
| L16 | B. dorei/B. vulgatus | B. massiliensis (2.39) | B. massiliensis |
| G12 | B. ovatus/B. xylanisolvens | B. ovatus (1.97) | B. ovatus/B. koorensis |
| G47 | B. ovatus/B. xylanisolvens | B. ovatus (2.06) | B. ovatus/B. koorensis |
| G58 | No identification | B. ovatus (1.80) | B. koorensis |
| G60 | No identification | B. nordii (1.96) | B. nordii |
| C22 | B. dorei | B. salyersiae (2.27) | B. salyersiae |
| HA66 | B. dorei/B. uniformis | B. coprocola (2.12) | B. coprocola |
| CEM30 | No identification | B. nordii (1.92) | B. nordii |
| CEM33 | B. fragilis | B. nordii (1.93) | B. nordii |
| Prevotella (n: 9) | |||
| G31 | No identification | P. heparinolytica (2.44) | P. heparinolytica |
| A35 | No identification | P. maculosa (2.03) | P. maculosa |
| A16 | No identification | P. bivia (2.25) | P. bivia |
| A21 | No identification | No identification | P. bivia |
| A33 | No identification | No identification | P. ihumii |
| CEM28 | No identification | No identification | P. oris |
| G75 | No identification | No identification | Prevotella sp. DNF 00663 |
| A58 | No identification | No identification | Alloprevotella rava |
| M60 | No identification | Prevotella sp. (2.00) | P. ihumii |
| Porphyromonas (n: 12) | |||
| L13 | P. asaccharolytica/P. uenonis | No identification | P. asaccharolytica |
| L19 | No identification | No identification | P. asaccharolytica |
| L50 | P. asaccharolytica/P. uenonis | No identification | P. asaccharolytica |
| A61 | No identification | P. asaccharolytica/P. uenonis (2.01) | P. asaccharolytica |
| A62 | No identification | P. asaccharolytica/P. uenonis (2.12) | P. asaccharolytica |
| CEM24 | No identification | No identification | P. bennonis |
| A68 | No identification | P. asaccharolytica/P. uenonis (1.88) | P. asaccharolytica |
| A69 | No identification | P. asaccharolytica/P. uenonis (1.83) | P. asaccharolytica |
| CEM25 | No identification | No identification | Porphyromonas sp. oral taxon 275 |
| A34 | No identification | No identification | Porphyromonadaceae sp. |
| A24 | No identification | P. endodontalis (2.11) | P. endodontalis |
| C27 | No identification | P. somerae (1.96) | P. somerae |
| Fusobacterium (n: 4) | |||
| L46 | F. nucleatum | F. nucleatum (1.53) | F. nucleatum |
| A8 | No identification | F. nucleatum (1.86) | F. nucleatum |
| L37 | F. gonidiaformans | F. gonidiaformans (1.59) | F. gonidiaformans |
| L36 | F. periodonticum | F. canifelinum (1.53) | F. canifelinum |
| Alistipes (n: 2) | |||
| CEM 20 | No identification | A. indistinctus (2.18) | A. indistinctus |
| CEM 22 | No identification | A. onderdonkii (2.08) | A. onderdonkii |
| Parabacteroides (n: 2) | |||
| L24 | No identification | No identification | P. faecis |
| Gram negative bacilli (GNB) | |||
| Bilophila (n: 2) | |||
| CEM 10 | No identification | B. wadsworthia (1.84) | B. wadsworthia |
| L42 | B. wadsworthia | B. wadsworthia (1.51) | B. wadsworthia |
| Pyramidobacter (n: 2) | |||
| CEM 29 | No identification | P. piscolens (2.33) | P. piscolens |
| CEM44 | No identification | P. piscolens (1.91) | P. piscolens |
| Gabonibacter (n: 1) | |||
| L43 | No identification | G. massiliensis (2.14) | G. massiliensis |
| Eikenella (n: 1) | |||
| A34 | No identification | No identification | Eikenella longinqua |
| Tidjanibacter (n: 1) | |||
| CEM 35 | No identification | No identification | T. massiliensis |
| Casaltella (n: 1) | |||
| G41 | No identification | No identification | C. massiliensis |
| Fenollaria (n: 2) | |||
| L31 | No identification | F. massiliensis (1.93) | F. massiliensis |
| M59 | No identification | F. massiliensis (1.97) | F. massiliensis |
| Gram positive bacilli (GPB) | |||
| Clostridium (n: 10) | |||
| G71 | No identification | Clostridium tunisiense (2.00) | Clostridium tunisiense |
| M50 | Clostridium sporogenes | No identification | Clostridium sporogenes |
| M55 | No identification | Clostridium subterminale (2.12) | Clostridium argentinense |
| G73 | No identification | No identification | Clostridium hydrogeniformans |
| L9 | No identification | Clostridium symbiosum (2.20) | Clostridium symbiosum |
| C9 | Clostridium sporogenes | Clostridium sp. 110324 (1.90) | Clostridium moniliforme/C. sardiniense |
| L10 | Clostridium clostridioforme | C. clostridioforme (1.89)/C. bolteae (1.80) | Enterocloster bolteae |
| L51 | No identification | C. aldenense (2.27) | Enterocloster aldenensis |
| G69 | No identification | C. celerecrescens (1.99)/C. sphenoides (1.85) | Lacrimispora celerecrescens |
| CEM21 | No identification | No identification | Lacrimispora amygdalina |
| Eggerthella (n: 3) | |||
| G8 | No identification | No identification | E. lenta |
| L33 | No identification | Eggerthella lenta (2.13) | E. lenta |
| CEM18 | No identification | Eggerthella lenta (1.89) | E. lenta |
| Paraeggerthella (n: 1) | |||
| G35 | No identification | No identification | P. hongkongensis |
| Gram positive bacilli (GPB) | |||
| Atopobium/Fannyhessea (n: 3) | |||
| G53 | No identification | No identification | Atopobium minutum |
| CEM 41 | No identification | Atopobium minutum (2.22) | Atopobium minutum |
| CEM 16 | No identification | Atopobium vaginae (1.72) | Fannyhessea vaginae |
| Bifidobacterium (n: 5) | |||
| L2 | No identification | B. scardovii (1.97) | B. scardovii |
| C48 | No identification | B. breve (1.98) | B. breve |
| CEM 13 | Bifidobacterium sp. | B. breve (2.06) | B. breve |
| CEM 14 | Bifidobacterium sp. | B. dentium (2.18) | B. dentium |
| G66 | Bifidobacterium sp. | B. dentium (1.76) | B. dentium |
| Slackia (n: 2) | |||
| L14 | No identification | S. exigua (2.07) | S. exigua |
| CEM 11 | No identification | S. exigua (2.24) | S. exigua |
| Solobacterium (n: 4) | |||
| CEM 4 | No identification | S. moorei (2.39) | S. moorei |
| CEM 5 | No identification | S. moorei (2.00) | S. moorei |
| CEM 6 | No identification | S. moorei (2.18) | S. moorei |
| CEM 7 | No identification | S. moorei (1.80) | S. moorei |
| Eubacterium (n: 1) | |||
| C37 | E. callanderi | No identification | E. limosum/callanderi |
| Tissierella (n: 2) | |||
| C11 | No identification | T. praeacuta (2.12) | T. praeacuta |
| G72 | No identification | T. praeacuta (2.08) | T. praeacuta |
| Moryella (n: 2) | |||
| CEM 27 | No identification | M. indoligenes (2.12) | M. indoligenes |
| CEM 45 | No identification | M. indoligenes (1.91) | M. indoligenes |
| Propionimicrobium (n: 1) | |||
| G70 | No identification | P. lymphophilum (1.95) | P. lymphophilum |
| Olsenella (n: 1) | |||
| G21 | No identification | O. uli (1.85) | O. uli |
| Filifactor (n: 1) | |||
| M51 | No identification | F. alocis (1.87) | F. alocis |
| Criibacterium (n: 1) | |||
| G64 | No identification | No identification | Criibacterium bergeronii |
| Gram positive cocci (GPC) | |||
| Peptoniphilus (n: 16) | |||
| A20 | No identification | P. duerdenii (2.36) | P. duerdenii |
| A38 | P. asaccharolyticus | P. indolicus (1.96)/P.harei (1.82) | P. harei |
| A40 | P. asaccharolyticus | P. indolicus (2.30)/P.harei (2.14) | P. harei |
| C56 | P. asaccharolyticus | P. indolicus (2.14)/P.harei (2.15) | P. harei |
| G11 | No identification | P. tyrrelliae (2.34)/P. senegalensis (2.30) | P. tyrrelliae |
| G17 | P. asaccharolyticus | P. harei (2.24)/P. indolicus (2.23) | P. harei |
| G37 | No identification | No identification | P. nemausensis |
| G40 | No identification | P. tyrrelliae (2.22)/P. indolicus (2.17) | P. tyrrelliae |
| G42 | P. asaccharolyticus | P. indolicus (2.37)/P. harei (2.17) | P. harei |
| G43 | P. asaccharolyticus | P. indolicus (2.02)/P. harei (1.85) | P. harei |
| G49 | P. asaccharolyticus | P. indolicus (2.13)/P. harei (2.06) | P. harei |
| G50 | No identification | P. rhinitidis (1.96) | P. lacydonensis |
| G51 | P. asaccharolyticus | P. indolicus (2.32)/P. harei (2.28) | P. harei |
| Anaerococcus (n: 11) | |||
| G10 | No identification | A. octavius (1.74) | A. octavius |
| G38 | No identification | A. murdochii (2.27) | A. murdochii |
| A54 | No identification | A. murdochii (1.83) | A. murdochii |
| G28 | No identification | No identification | A. mediterraneensis |
| G44 | No identification | A. octavius (2.02) | A. octavius |
| G49 | No identification | A. nagyae (2.09) | A. nagyae |
| A63 | No identification | No identification | A. urinomassiliensis |
| A64 | No identification | No identification | A. provencensis |
| C55 | A. vaginalis | No identification | A. jeddahensis |
| G55 | No identification | A. octavius (2.12) | A. octavius |
| G56 | No identification | A. hydrogenalis (1.91) | A. hydrogenalis |
| Finegoldia (n: 1) | |||
| C54 | F. magna | F. magna (1.68) | F. magna |
| Peptostreptococcus (n: 3) | |||
| G5 | No identification | No identification | P. stomatis |
| G15 | No identification | No identification | P. stomatis |
| G24 | No identification | P. stomatis (1.90) | P. stomatis |
| Parvimonas (n: 1) | |||
| M48 | No identification | Parvimonas micra (1.63) | Parvimonas sp. |
| Staphylococcus (n: 1) | |||
| C62 | No identification | S. saccharolyticus (1.94) | S. saccharolyticus |
| Murdochiella (n: 2) | |||
| G30 | No identification | No identification | Murdochiella vaginalis |
| C61 | No identification | M. asaccharolytica (2.10) | M. asaccharolytica |
| Lagierella (n: 1) | |||
| G46 | No identification | No identification | L. massiliensis |
| Gram positive cocci (GPC) | |||
| Fastidiosipila (n: 2) | |||
| CEM36 | No identification | F. sanguinis (1.98) | F. sanguinis |
| CEM37 | No identification | F. sanguinis (1.89) | F. sanguinis |
| Gram negative cocci (GNC) | |||
| Acidaminicoccus (n: 3) | |||
| CEM1 | No identification | A. intestini (2.36) | A. intestini |
| CEM2 | No identification | A. intestini (2.38) | A. intestini |
| L45 | No identification | No identification | A. intestini |
| Negativicoccus (n: 1) | |||
| CEM38 | No identification | N. succinivorans (1.71) | N. succinivorans |
The isolates that were not identified or that showed discrepancies were further analyzed by 16S rRNA gene sequencing (Table 4). In 7 isolates (L5, G47, G60, C22, HA66, CEM30 and CEM33) 16S rRNA sequencing and Bruker Biotyper identification were concordant. Inconsistency was observed in the identification of G58 isolate that was identified as Bacteroides koorensis by its 16S rRNA sequence, but Bruker Biotyper assigned it as Bacteroides ovatus.
The analysis of the 16S rRNA gene sequence was useful to correct the discrepancies in 8 out of 10 Bacteroides isolates; however, in 2 isolates, it was observed that there was >99% sequence identity for B. ovatus and B. koorensis. In 2017 Shin et al. described a new species, B. koorensis, from isolates recovered in human feces, which displayed relatedness with B. ovatus and B. xylanisolvens64.
It is important to highlight that an additional note from the Vitek MS database (V3.2.0) revealed that B. ovatus is grouped with a diagonal line with B. xylanisolvens; however, B. xylanisolvens is not included in the Bruker Biotyper database yet. Therefore, an alternative could be to inform the complex B. ovatus/B. xylanisolvens when B. ovatus is present in clinical environments. B. ovatus/B. xylanisolvens showed a close phylogenetic relationship and they evidence biochemical similarity15. Undoubtedly, to enable a more accurate identification, the databases should be expanded.
Nevertheless, the identification accuracy of MALDI-TOF MS for Bacteroides species was studied and proved to be superior to biochemical testing15,46.
Bacteroides is one of the most common and well-known genera that contains numerous species that can be found in the human gut microbiome. Other related genus such as Parabacteroides, Alistipes and Odoribacter belonging to the same Bacteroidales order, are relatively new and can also be relevant in some infectious diseases31,35,49,59. Two isolates of Alistipes genus (Alistipesindistinctus and Alistipes onderdonkii) were correctly identified only by Bruker Biotyper. This identification was confirmed by 16S rRNA gene sequencing (Table 4).
Alistipes genus includes 10 species validly published (https://lpsn.dsmz.de/search?word=alistipes). Currently, 5 species are represented in the Bruker Biotyper database (Alistipes finegoldii, A. onderdonkii, Alistipes shahii, A. indistinctus and Alistipes putredinis) and only one species is represented in the Vitek MS database (A. finegoldii). Parker et al. mentioned that the identification in clinical samples is often underestimated and that it would be necessary to update databases49.
Odoribacter genus includes two species validly published in human samples, Odoribacter laneus and Odoribacter splanchnicus. The two isolates of O. splanchnicus included in this work were correctly identified by both MALDI-TOF MS systems (Tables 2 and 3)31.
With regard to Parabacteroides spp., 4/6 isolates included in this study were identified at the species level by both MALDI-TOF MS systems. The remaining two unidentified isolates (L24 and C57) corresponded to Parabacteroides faecis and Parabacteroides chongii as per their 16S rRNA sequence. None of these species are included in the MALDI-TOF MS databases. P. faecis was described in 2015 as a new species isolated from human feces and closest related (96% closest similarity) to Parabacteroides gordonii based on 16S rRNA sequence analysis58. P. chongii is a recently described species and the 16S rRNA gene sequence is closely related to P. faecis (97.3% identity), P. gordonii (96.6% identity), and P. goldsteinii (95.7% identity)35.
Both MALDI-TOF MS systems presented troublesome identification when isolates of Prevotella spp. and Porphyromonas spp. were analyzed. The reason could be their pigmented nature that hindered the quality of spectra obtained, and the inadequate number of spectra of the represented species13,54,81.
Accurate identification of Prevotella isolates plays a critical role in the success of the treatments, especially since the antibiotic susceptibility profile differs between species73. In 2012, Wybo et al. studied 102 clinical Prevotella isolates and only 63% were identified at the species level80. In a subsequent study, the expansion of the commercial database increased the correct identification of the species reaching 89%27. In the present study, the Vitek MS system identified 20/28 isolates (71.4%), and Bruker Biotyper 23/28 isolates (82.1%) (Table 2). The species identified by both systems were: Prevotella buccae, Prevotella baroniae, Prevotella nigrescens, Prevotella timonensis, Prevotella nanciensis, Prevotella denticola and Prevotella bergensis.
Nine isolates were sequenced due to discrepancies or lack of identification between the MALDI-TOF MS systems (Table 4). Three isolates were identified by Bruker Biotyper and confirmed by 16S rRNA sequencing as Prevotella heparinolytica, Prevotella maculosa and Prevotella bivia. Among the isolates that could not be identified by any of the MALDI-TOF MS systems not even at the genus level, one isolate corresponded to Prevotella ihumii, and another one to Alloprevotella rava, which are currently missing or there is only one MSP in the databases. Anani et al. recently described the species P. ihumii, a bacterium isolated from a stool specimen of a healthy woman whose reference MSP was imported into their own database (http://www.mediterranee-infection.com/article.php?larub=280&titer=urms-database)3. Only Bruker Biotyper identified the M60 isolate at the genus level, which corresponded to P. ihumii (Table 4).
However, some species that are well-represented in the database such as Prevotella oris and P. bivia were not identified.
One isolate could neither be identified by the MALDI-TOF MS sytems nor by 16S rRNA secquencing, and only displayed identity with Prevotella sp. DNF 00663 (Accession Number KF280297.1). More studies should be performed to reach the species level, as it could correspond to a new species.
In the identification of Porphyromonas spp., the performance of the MALDI-TOF MS systems was different. Using the Vitek MS system, only 12 out of 22 isolates were identified. Nowadays, the database includes only three species out of 25 listed in the List of Prokaryotic names with Standing in Nomenclature (LPSN). According to Vitek MS, 12/22 isolates studied, belonged to Porphyromonas gingivalis and Porphyromonas asaccharolytica/Porphyromonas uenonis, but further analysis evidenced that they were distributed among six different species. The analysis of the 16S rRNA sequences for five isolates from this complex (L19, A61, A62, A68 y A69) showed the highest identity with P. asaccharolytica (Table 4). Moreover, the analysis of the 16S rRNA sequences was needed to identify 3/22 isolates that corresponded to Porphyromonas endodontalis, Porphyromonas somerae and Porphyromonas bennonis. Of these, P. endodontalis and P. somerae were correctly identified using the Bruker Biotyper system. Although P. bennonis is represented in the Bruker Biotyper database, the isolate CEM24 was not identified. It should be noted that there is only one MSP of this species. Seng et al. previously reported that poor bacterial identification is mostly due to an insufficient number of spectra in the database63. However, for other several species, low database sampling does not interfere with a good identification level36. Isolates L13, L19 and L50 corresponding to P. asaccharolytica by 16S rRNA sequences displayed a very low score (<1.5) for P. asaccharolytica in the top ten identification using the Bruker Biotyper system (Table 3). On the other hand, isolates CEM25 and A34 that were not identified by the MALDI-TOF MS systems corresponded to unidentifiable taxa by their 16S rRNA sequence analysis, being suggestive of a new species; however, more studies are necessary to classify these isolates.
The literature reported that the identification of Porphyromonas spp. by MALDI-TOF MS exhibited problems at both the species and genus levels. Vega-Castaño et al. showed that none of the 10 isolates of P. asaccharolytica studied could be identified by MALDI-TOF MS, although a different database version was used (version 2.0)72. In another study of Rodríguez-Sánchez et al., none of the two isolates (P. somerae and P. asacharolytica/P. uenonis) could be identified by MALDI-TOF MS57. In a recent analysis, Alcalá et al. included 14 isolates and only P. somerae and P. gingivalis could reach the identification at the species level by MALDI-TOF MS2. The difference in the identification performance by both authors could be explained by the number of MSP entries in the database (Rodríguez-Sánchez et al., 5627 MSP entries vs Alcalá et al., 9234 MSP entries).
Among the rare species detected in this order, one isolate was identified as Gabonibacter massiliensis and the other corresponded to Tidjanibacter massiliensis.
G. massiliensis was correctly identified by the Bruker Biotyper system and confirmed by the 16S rRNA analysis. However, both MALDI-TOF MS failed to identify T. massiliensis and only the 16S rRNA analysis allowed it. According to LPSN, the Tidjanibacter genus belongs to the Rikenellaceae family and contains only one species published in 2017 from the human colon, which was phylogenetically closest to Alistipes putredinis (divergence of >5%)43. MALDI-TOF MS MSP of T. massiliensis is available online (http://www. mediterranee infection.com/article.php?laref=256&titre=urmsdatabase); however, it was not included in the Vitek and Bruker Biotyper databases.
- ∘
Class Fusobacteriia. Order Fusobacteriales
Identification at the species level was achieved in 14/16 and 13/16 isolates using Vitek MS and Bruker Biotyper, respectively. One out of two isolates, L36, which could not be identified at the species level by Vitek MS corresponded to Fusobacterium canifelinum, a species not included in its database (Tables 2 and 3). Interestingly, the L36 isolate showed the top ten identification score with F. canifelinum using Bruker Biotyper; however, the species level could not be assigned since the score was <1.7. The other two isolates did not achieve the proper score for species level identification using Bruker Biotyper: Fusobacterium nucleatum, score 1.53, and Fusobacterium gonidiaformans, score 1.59. However, these species were confirmed by the 16S rRNA analysis (Table 4).
The difficulty of identifying Fusobacterium spp. by MALDI-TOF MS has been previously reported. On the other hand, for one of the most common species F. nucleatum, the 16S rRNA sequences showed divergences between 0.6% and 1.9%, defining it as a highly heterogeneous species2,23,63.
- ∘
Class Epsilonproteobacteria. Order Camkpylobacterales and
- ∘
Class Negativicutes. Order Veillonellales
The identification of Campylobacter spp. (n: 11) and Dialister spp. (n: 5) was correctly achieved by both MALDI-TOF MS systems.
- ∘
Class Deltaproteobacteria. Order Desulfovibrionales
Two isolates corresponded to the genus Bilophila using 16S rRNA sequencing, which were correctly identified at the genus level using the Bruker Biotyper system, but only one at the species level, as the score was <1.7. Using the Vitek MS system, one isolate was assigned as Bilophila wadsworthia, but the remaining one was not identified. In a recent study, Alcalá et al. also reported reliable identification for this genus by MALDI-TOF MS2. One isolate recognized as Desulfovibrio desulfuricans was correctly identified by both systems.
- ∘
Rare species
Among GNB, only the Bruker Biotyper system correctly identified isolates that corresponded to Pyramidobacter piscolens (n: 2) which belongs to class Synergistia and order Synergistales19, as well as Fenollaria massiliensis (n: 2) isolates from class Clostridia and order Eubacteriales. Boiten et al. have pointed out the importance of adding more spectra of less common species such as F. massiliensis to the MALDI-TOF MS databases to gain insight into their clinical relevance10.
Two isolates of GNB have only been identified by the 16S rRNA analysis. One isolate corresponded to class Betaproteobacteria and order Neisseriales and has been identified as Eikenella longinqua. Until 2020, the genus Eikenella contained a single species, Eikenella corrodens, which belongs to the HACEK (Haemophilus, Aggregatibacter, Cardiobacterium, Eikenella and Kingella) group considered as miscellaneous or fastidious gram negative facultative anaerobic bacteria. Recently, the emendation of this genus included three new species of strict anaerobic GNB: E. longinqua, Eikenella halliae and Eikenella exigua9.
The other one was identified as Casaltella massiliensis. This species was mentioned by La Scola et al. in an infected lipectomy as a small gram negative bacillus, indole positive, whose sequence was introduced in Genbank in 2017 (Accession Number HM587320)36. Although, C. massiliensis is not validated nor included in LPSN.
- -
Identification of gram positive bacilli (GPB) isolates
The GPB corresponded to five classes and eight orders. Out of 110 GPB isolates, 75 (68.2%) and 80 (72.7%) were identified at the species and genus levels, respectively, using the Vitek MS system. Using the Bruker Biotyper system, high performances were observed, as 98 (89.1%) and 101 (91.8%) of GBP were identified at the species and genus levels, respectively. The differences in performances for species and genus, were statistically significant for both systems, with respective p-values of 0.0003 and 0.0004.
Thirty-three isolates (30%) were not identified at the species level and two isolates (1.8%) were misidentified using the Vitek MS system. Meanwhile, 12 isolates (10.9%) were not identified and there were no misidentified isolates using the Bruker Biotyper system (Tables 2 and 3). In GPB identification, the main trouble was related to the lack of spectra in the databases, which was more evident using the Vitek MS system.
- ∘
Class Clostridia. Order Eubacteriales.
- ∘
Family Clostridiaceae
With regard to Clostridium spp., four isolates were not identified using the Vitek MS system that corresponded to Clostridium tunisiense, Clostridium symbiosum, Clostridium argentinense, and Clostridium hydrogeniformans, which have not been included in the database. Isolate M55 identified as C. argentinense corresponded to Clostridium subterminale by the Bruker Biotyper system. Suen et al. described this species as the first toxigenic strain isolated from Argentinian soil, and they mentioned it as a genetically homogeneous group of strains previously identified as C. subterminale67. Therefore, identification using the Bruker Biotyper system was considered correct to the species level for this isolate. On the other hand, the Bruker Biotyper system failed to identify C. hydrogeniformans, since its database does not include it. Additionally, this system failed in the identification of one isolate of Clostridium sporogenes despite being part of the database. C. hydrogeniformans as well as C. tunisiense were described from chlorinated solvent-contaminated groundwater and olive mill wastewater, respectively, but they were not described in human samples11,68.
Both systems were efficient in the identification of Clostridioides difficile, the most common nosocomial pathogen in antibiotic-related diarrhea into health care facilities16.
Furthermore, the five Paeniclostridium sordellii (former Clostridium sordellii) isolates were correctly identified by the two platforms.
The data obtained from the whole genome sequencing entailed taxonomic changes in the genus Clostridium, which was reclassified into two separate clades. One clade which includes Clostridium clostridioforme, Clostridium aldenense and Clostridium bolteae, which was reclassified as Enterocloster gen. nov., and another clade that comprises Clostridium sphenoides, Clostridium amygdalinum and Clostridium celerecrescens, which was reclassified as Lacrimispora gen. nov.28. Isolate G69, identified by 16S rRNA sequencing as Lacrimispora celerecrescens could not be identified by the Vitek MS system, since it is not included in the database. Instead, the Bruker Biotyper system identified it as C. celerecrescens/C. sphenoides. A unique identification could not be assigned because the difference in scores between them did not exceed 10%. These two species display the closest relationship between them (98.1% identity). In a recent case of chronic osteomyelitis by C. sphenoides, several discrepancies in identification using MALDI-TOF MS and 16S rRNA sequencing were observed. Whole genome sequencing was necessary to solve it51. In summary, we suggest considering these two species as a complex when MALDI-TOF MS is used as the identification method.
Neither the Vitek MS nor Bruker Biotyper databases included the species Lacrimispora amygdalina (or C. amygdalinum); therefore, they failed in the identification at the species or genus levels.
With respect to species that were isolated in small numbers in this study, one isolate of Robinsoniella peoriensis was correctly identified by both MALDI-TOF MS, since this species is included in both databases. Only a few cases were published in which R. peoriensis was identified as the cause of an infection. Schrottner et al. described the detection of R. peoriensis in multiple bone samples of a trauma patient. The bacterium could only be identified using 16S rRNA sequencing, since the results by MALDI-TOF MS system gave a score below 1.7, which could not be considered as secure identification at the species level. They used the Bruker Biotyper database which contains 7854 reference spectra (version 8.0), which is an older version than the one that was used in the present study62.
Veloo et al. recently validated the use of the Bruker MALDI-TOF MS database, by using a large set of anaerobic strains isolated from human clinical specimens where 4 isolates of C. aerofaciens were identified with score ≥2.075. In contrast to our study, Lee et al. could not identify the only one isolate of C. aerofaciens using the Vitek MS system, since they used an older version38.
Two isolates of Eubacterium spp. were included in this study and were both identified by the Vitek MS system, but only one was identified by the Bruker Biotyper system. When 16S rRNA sequencing was performed, in the C37 isolate (Table 4) we observed that both species, Eubacterium callanderi and Eubacterium limosum, could not be differentiated using this gene sequence, as they share more than 99% identity. Therefore, in this case, the correct identification might be considered as E. callanderi/E. limosum complex. Li et al. reported that the identification accuracy of MALDI-TOF MS was 57% for Eubacterium spp.40.
The genera Moryella and Filifactor are only included in the Bruker Biotyper database, therefore, the species Moryella indoligenes (n: 2 isolates) and Filifactor alocis (n: 1) were only identified using this system and later confirmed by 16S rRNA gene sequencing.
An isolate that could not be identified by either of the two MALDI-TOF MS systems, was identified by 16S rRNA gene sequencing as Criibacterium bergeronii. This novel species was recently described in 2021 from a vaginal sample of a woman with bacterial vaginosis, and has not been included in these databases yet42.
- ∘
Class Actinomycetes.
- ∘
Order Propionibacteriales
All the isolates (n: 13) that belong to Cutibacterium spp., formerly Propionibacterium spp., were correctly identified by both MALDI-TOF MS systems. Traditionally, the species within this genus were grouped as either classical or cutaneous propionibacteria. This group that used to comprise the species Propionibacterium. Propionibacterium acnes, Propionibacterium avidum and Propionibacterium granulosum were accommodated into the genus Cutibacterium gen. nov. by Scholz and Killian61. Peel et al., highlighted the ability of MALDI-TOF MS to quickly identify this kind of gram positive bacilli, meaning an important tool to assess their clinical significance50.
Only one isolate from a patient with a urinary tract infection was identified as Propionimicrobium lymphophylum by the Bruker Biotyper system and confirmed with 16S rRNA sequencing; however, it could not be identified by Vitek MS, as it is not present into the database. This genus was created to accommodate P. lymphophylum, a species that is a rarely encountered anaerobic gram positive non-spore forming rod that might be an emergent uropathogen66,78.
- ∘
Order Bifidobacteriales
Regarding the order Bifidobacteriales,Bifidobacterium, is one of the genera that includes the largest number of species, either facultative or strict anaerobic. All isolates, Bifidobacterium dentium (n: 2), Bifidobacterium breve (n: 2) and Bifidobacterium scardovii (n: 1), were correctly identified at the species level by the Bruker Biotyper system. However, when using Vitek MS, they could only be identified at the genus level as Bifidobacterium spp. Moreover, the Vitek MS database manual mentions that for the species Bifidobacterium adolescentis, Bifidobacterium bifidum, B. breve, B. dentium, and Bifidobacterium longum, the system will identify them as Bifidobacterium spp. Only one B. scardovii isolate could not be identified, although this species is included in the Vitek MS database.
- ∘
Order Actinomycetales
The order Actinomycetales does not include strict anaerobic species, but since they are fastidious and slow growing species in the aerobic atmosphere, they were included in our study. Actinomyces (n: 3) and Actinotignum (n: 1) isolates were correctly identified by both MALDI-TOF MS systems.
Barberis et al. demonstrated high efficacy in the identification at the species level in these bacteria using Bruker Biotyper MALDI-TOF MS6.
- ∘
Class Coriobacteriia.
- ∘
Order Eggerthellales
Eight isolates of Eggerthella lenta (first described in 1935 as Eubacterium lentum by Arnold Eggerth) were included20. This bacterium was characterized in more detail through genetic analysis in 1999, placing it in its distinct genus76. Three isolates could not be identified by the Vitek MS system, and two isolates could not be identified by the Bruker Biotyper system, although this species is included in both databases. Conversely, Alcalá et al. recently reported, in a four-year experience in MALDI-TOF MS identification of anaerobic bacteria, one of the largest studies about E. lenta isolates (n: 71), and most of them (n: 66) were correctly identified at the species level2.
The complete genomic sequence of a relatively new closely related species, Paraeggerthella hongkongensis, was published in 2009, but it was not included in the MALDI-TOF MS systems79. Thus, the only isolate included in our study was not identified. This species was also involved in bacteremia cases, similar to E. lenta37. Five isolates of Slackiaexigua were included and were correctly identified at the species level by Bruker Biotyper, only three of them were identified at the species level using the Vitek MS system. Li et al. observed that the identification accuracy of MALDI-TOF MS against the Slackia genus was 83%40.
- ∘
Order Coriobacteriales
Eight isolates of Atopobium spp. were included in this study. The two isolates that were identified as Atopobium minutum by 16S rRNA gene sequencing were not identified by the Vitek MS system, as this species is not included in the database, and only one isolate was correctly identified at the species level by the Bruker Biotyper system. The five isolates of Atopobium rimae (n: 2) and Atopobium parvulum (n: 3) agreed in the identification by both systems. On the other hand, Atopobium vaginae (n: 1) was correctly identified only by the Bruker Biotyper system even though this species is included in Vitek MS system database. Recently Nouioui et al. reported changes based on the genome taxonomic classification that placed the species A. rimae and A. parvulum into the genus Lancefieldella, and A. vaginae into the genus Fannyhessea being the correct names Lancefieldella rimae, Lancefieldella parvula and Fannyhessea vaginae, respectively47.
With regard to species that were isolated in small numbers in this study, one isolate of Collinsella aerofaciens was correctly identified by both MALDI-TOF MS systems, as this species is included in both databases.
The Olsenella genus is only included in the Bruker Biotyper database; therefore, Olsenella uli (n: 1) included in this study, was only identified at the species level using this system and then confirmed by 16S rRNA sequencing.
- ∘
Class Erysipelotrichia. Order Erysipelothricales
Isolates from the genera Eggerthia and Solobacterium were analyzed. We observed that three isolates of Eggerthia catenaformis, were correctly identified by both MALDI-TOF MS systems, as these species are included in both databases. E. catenaformis (formerly known as Lactobacillus catenaformis) is a member of the human fecal microbiota, rarely associated with human infections and reassigned to the Eggerthia genus by Salvetti et al. in 201160. Foronda et al. reported the second case of bacteremia due to this organism, which could be identified using MALDI-TOF MS25.
Four isolates of Solobacterium moorei, the only species included in the genus, were evaluated. As we observed with other anaerobes, S. moorei is an example of the differences that exist between currently available databases, as it is absent from the Vitek MS database. Alauzet et al. carried out a retrospective analysis of 27 cases of infection involving S. moorei that had to be identified by 16S rRNA gene sequencing because they used the Vitek MS system1.
- ∘
Class Tissierellia. Order Tissierellales
The Tissierella genus is included in the Bruker Biotyper database, but not in that of the Vitek MS. Therefore, the two Tissierella praeacuta isolates included in this study were only identified at the species level using the Bruker Biotyper system, and then confirmed by 16S rRNA sequencing.
Veloo et al. validated the Bruker database optimized for anaerobic bacteria, including strains of species less commonly encountered in human infections. They observed that the addition of more spectra optimized the database and improved identification with higher confidence75. Undoubtedly, Vitek MS should expand its database to include less frequently isolated species as this would allow to gain insight into the clinical relevance of these less common anaerobic bacteria.
- -
Identification of gram positive cocci (GPC) isolates
Most of the GPC belong to the class Clostridia, order Eubacteriales with the genera Peptoniphilus spp., Anaerococcus spp., Finegoldia sp., Peptostreptococcus spp., Parvimonas sp., Murdochiella spp., Fastidiosipila sp., Ruminococcus sp., and Lagierella sp. Additionally, two isolates of class Bacteria, order Caryophanales were included. Eighty-eight percent (66/75) and 82.7% (62/75); 65.3% (49/75) and 54.7% (41/75) of the GPC were identified at the genus level and species level using the Bruker Biotyper system and the Vitek MS system, respectively. The differences in the performances of both systems for genus and species, were statistically significant, with respective p-values of 0.002 and 0.0004.
- ∘
Class Clostridia. Order Eubacteriales.
- ∘
Family Peptoniphilaceae
In a recent study conducted by our group, the identification of 18 isolates of Peptoniphilus spp. was analyzed and it was demonstrated that the performance of the Bruker Biotyper system outperformed the Vitek MS system for this genus7. These results were included in this study (Tables 2 and 4).
Finegoldia is a genus represented by only one species, Finegoldia magna (https://lpsn.dsmz.de/genus/finegoldia). It is part of the human normal microbiota; however, it is considered one of the most pathogenic species among anaerobic gram positive cocci, as it displays a variety of virulence factors41. All the isolates included in this study (n: 17) were correctly identified at the species level using the Vitek MS system. Bruker Biotyper identified 16 isolates at the species level, as one isolate gave a score <1.7. Alcalá et al. in an extensive study of the identification of anaerobes by MALDI-TOF MS (Bruker) that included 299 isolates of Finegoldia magna, showed that most of the isolates (n: 290) were identified with scores ≥2.0, 95 isolates with scores 1.99–1.7, and 13 isolates with scores 1.6–1.692. Therefore, we consider that not only the MALDI-TOF MS is a great tool for the identification of this species, but also lower scores than those recommended by the manufacturer could be considered when the top ten assigns F. magna.
Regarding Anaerococcus spp., we observed that it was one of the CGP that presented more difficulties in the identification at the species level. At present, nine species are represented in the Bruker Biotyper database (version 10.0) (Anaerococcus degeneri, Anaerococcus hydrogenalis, Anaerococcus lactolyticus, Anaerococcus murdochii, Anaerococcus nagyae, Anaerococcus octavius, Anaerococcus prevotii, Anaerococcus tetradius, Anaerococcus vaginalis), and only three species are represented in the Vitek MS database (A. prevotii, A. tetradius and A. vaginalis). Consequently, all isolates included in this study that belong to the species A. prevotii, A. tetradius and A. vaginalis were correctly identified by the Vitek MS system. However, the other isolates included in the study that belong to species not included in the Vitek MS database (n: 10) were not identified or misidentified. One isolate identified by 16S rRNA gene sequencing as Anaerococcus jeddhahensis was misidentified as A. vaginalis. Conversely, using the Bruker Biotyper system, six isolates could not be identified, but none were misidentified. These isolates were identified by 16S rRNA gene sequencing as Anaerococcus urinomassiliensis, A. jeddahensis, Anaerococcus mediterraneensis, Anaerococcus provencensis18,44,48. None of these recently described species have been included in other studies evaluating the performance of MALDI-TOF MS2. This highlights the importance of including new species in the database to accurately assess their clinical impact on human infections.
After the reclassification of the species of Peptostreptococcus into other several genera, the remaining members were Peptostreptococcus anaerobius21,74 and then, Peptostreptococcus stomatis, both isolated from human samples. These two species are included in the Bruker Biotyper database but only one species, P. anaerobius, is included in the Vitek MS database. As expected, all P. anaerobius isolates included in this study (n: 5) were correctly identified by both MALDI-TOF MS systems, but none of the P. stomatis isolates (n: 3) could be identified using the Vitek MS system. One of three P. stomatis was identified only at the genus level using the Bruker system, as the score obtained was <1.7. In the study by Alcalá et al., the only species included was P. anaerobius and only 2/36 isolates showed a score <1.72.
It is known that only one species is included in the genus Parvimonas. Parvimonasmicra has a long-standing presence in nomenclature and its role in human health and disease has been studied to some extent70. Recently a new species, Parvimonas parva was described. All but one of the isolates included in this study (n: 7) were correctly identified at the species level for both MALDI-TOF MS systems. Alcalá et al. showed, in one of the largest number of isolates (n: 255) included in the evaluation of MALDI-TOF MS performance, that more than 99% were correctly identified at the species level2.
Two isolates of Murdochiella spp. were also included in this study. One of them was identified as Murdochiella asaccharolytica by the Bruker Biotyper system and confirmed by 16S rRNA sequencing. It was not identified by the Vitek MS system, as it is not present in its database. The other isolate was not identified by any MALDI-TOF MS system. Its 16S rRNA analysis corresponded to Murdochiellavaginalis (Accession Number LT576397.1).
Fastidiosipila sanguinis, the only species of the genus Fastidiosipila, is currently not included in the MALDI-TOF database for the Vitek MS system, but has just been included in the Bruker Biotyper database revision no. 10. For the two isolates included in this study, identification using 16S rRNA sequencing resulted in species identification with more than 99.9%. So far, in the literature, the few published cases that involved F. sanguinis infection reported the same results22,29.
For species of which only one strain was encountered, we found one isolate of Ruminococcus gnavus. This species is represented in both databases and therefore was correctly identified by both MALDI-TOF MS systems. Fontanals et al. were able to identify R. gnavus using the Bruker Biotyper system with a score >2.0 from blood culture24.
Hansen et al. initially identified two R. gnavus bloodstream infections with scores of 1.675 and 1.723 using the Bruker Biotyper software v.3.0 and later obtained scores of 2.181 and 2.124 using software v.3.1. This underlines the importance of keeping databases up to date30.
Conversely, one isolate of Lagierella massiliensis was only identified by 16S rRNA sequencing, as it is not represented in the databases. The MALDI-TOF MS MSP of L. massiliensis is only available at http://www.mediterraneeinfection.com/article.php?laref=256&titreurms-database71.
- ∘
Class Bacilli. Order Caryophanales
Staphylococcus saccharolyticus is a rarely encountered coagulase-negative cocci and is the only strictly anaerobic species within the genus Staphylococcus77. The two isolates included in this study were correctly identified at the species level using the Bruker Biotyper system; however, although this species is also included in the Vitek database, only one of the isolates was identified by Vitek MS.
- -
Identification of gram negative cocci (GNC) isolates
The GNC corresponded to class Negativicutes and the orders Acidaminococcales and Veillonellales, which include the most frequently described genera in the literature.
Regarding the order Veillonellales, two isolates of Veillonella atypica and one isolate of Veillonella parvula were correctly identified at the species level by both MALDI-TOF MS systems. Accordingly, Alcalá et al. showed that 99.5% of the GNC included in their study, mostly Veillonella species, were correctly identified at the species level2. However, Veloo et al. have pointed out that several species of Veillonella, such as Veillonella dispar, V. parvula, Veillonella denticariosi and Veillonela rogosae, are difficult to separate by MALDI-TOF MS and 16S rRNA gene sequencing; therefore, any of these four species should be named as Veillonella spp.75. Further research into the identification of Veillonella using MALDI-TOF MS is necessary.
Negativicoccus spp. could not be identified by Vitek MS, as these species are not included in the database. The only included isolate of Negativicoccus succinivorans was correctly identified by Bruker Biotyper.
With regard to the order Acidaminococcales, neither of the Acidaminococcus spp. could be identified by Vitek MS, since they are not included into its database. Two out of three Acidaminococcus intestini isolates were correctly identified at species level by Bruker Biotyper, and all of them were confirmed by 16S rRNA gene sequencing.
ConclusionsMALDI-TOF MS systems speed up microbial identification and are especially effective for slow-growing microorganisms, such as anaerobic bacteria, which are difficult to identify by traditional methods.
When assessing GNBs, isolates of Bacteroides spp., the most frequent GNB found in clinical laboratories, were correctly identified by both systems. However, the GNBs that were not reliably identified mainly corresponded to the Porphyromonas genus, followed by the Prevotella genus. The species most frequently isolated within GPBs, Clostridium/Clostridioides, were correctly identified by both systems. However, Bruker showed advantages in specific species within this genus, as they were absent in the Vitek MS database. Concerning CGPs, Anaerococcus spp. was the genus that presented more difficulties in the identification at the species level for both systems. Misidentification observed in Peptoniphilus spp. by the Vitek MS system was mainly attributed to its failure to identify P. harei.
In summary, the Bruker system was more accurate than the Vitek system. In order to be truly effective, it is essential to update the databases of both systems by increasing the number of each MSP within their platforms.
FundingThis work was partially supported, equally by Becton Dickinson (BD) and bioMèrieux, Argentina.
Conflict of interestNone declared.