The family Dicistroviridae comprises three genera and about twenty species of RNA virus, most of them with health or agricultural importance. The Triatoma virus (TrV) is the only entomopathogenic virus identified in triatomine bugs up to the present. TrV replicates within the intestinal epithelial cells, causing high mortality rate and delayed development of the molt of these bugs. TrV has been proposed as a biological control agent for vectors of Chagas disease. Viral particles were purified from feces of 1, 5 and 10 insects from an experimental colony of Triatoma infestans infected with TrV. Viral concentration and infectivity were corroborated using polyacrylamide gels and RT-PCR, respectively. In this work we report a method of viral purification that allows to reduce necessary reagents and time, using a very small amount of fecal matter.
La familia Dicistroviridae está compuesta por tres géneros y casi una veintena de especies de virus ARN, la mayoría de ellas de importancia sanitaria o agrícola. Triatoma virus (TrV) es el único virus entomopatógeno identificado en triatominos hasta el momento. El TrV se replica en las células del epitelio intestinal; ello provoca una alta tasa de mortalidad y retraso en el desarrollo de la muda del insecto. Se ha propuesto la utilización de TrV como agente de control biológico para vectores de la enfermedad de Chagas. Las partículas virales fueron purificadas a partir de materia fecal de 1, 5 y 10 insectos obtenidos de una colonia experimental infectada con TrV de Triatoma infestans y se corroboró su concentración viral e infectividad mediante geles de poliacrilamida y RT-PCR, respectivamente. En este trabajo se reporta un método de purificación viral que permite la reducción de los reactivos y del tiempo necesario para lograr dicha purificación, partiendo de una mínima cantidad de materia fecal.
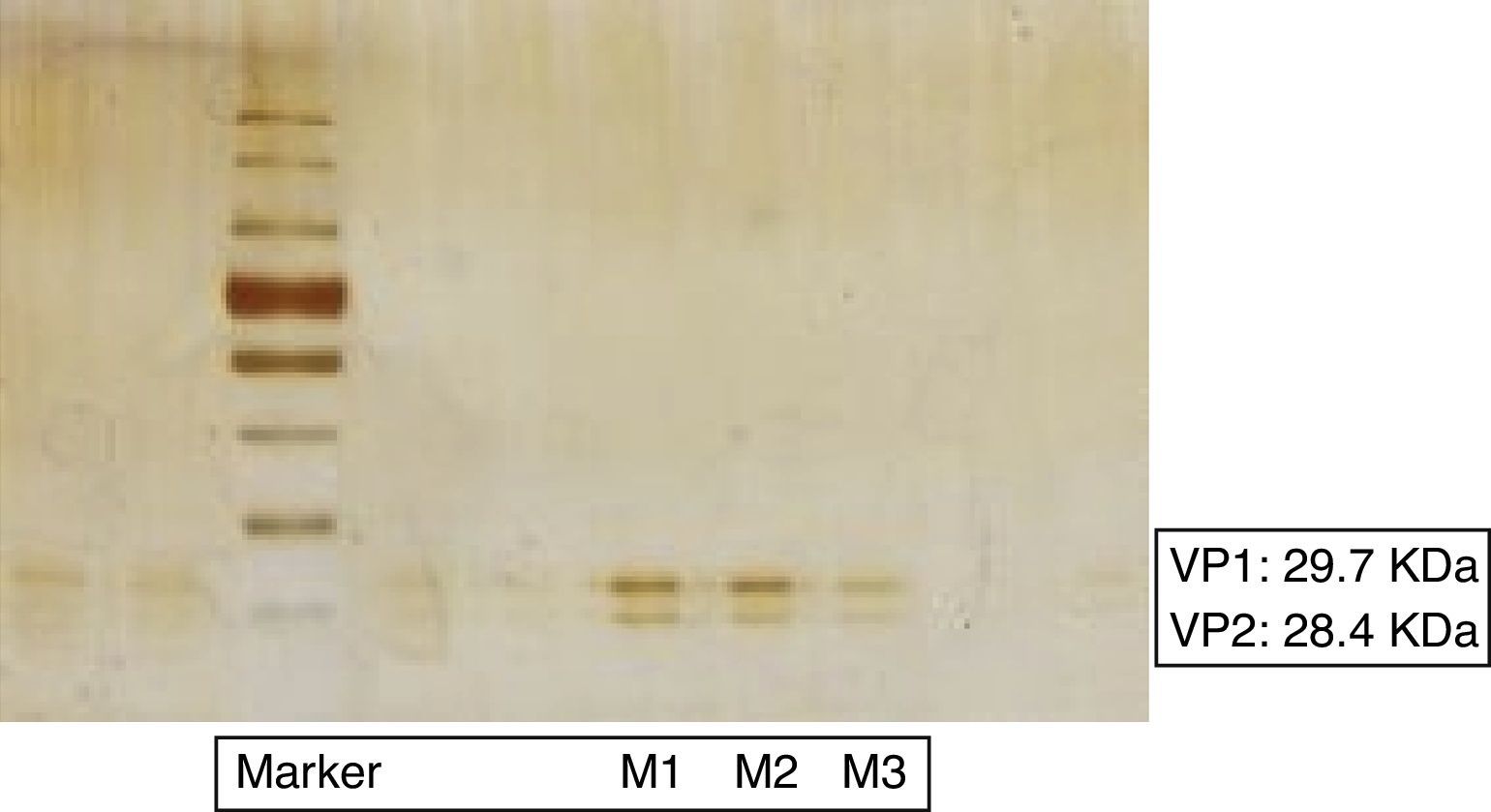
The family Dicistroviridae includes three genera: Cripavirus, Aparavirus and Triatovirus, with almost twenty species, most of which have sanitary or agricultural importance3,6. The name of the family refers to the two open reading frames or cistrons (ORFs) that codify its viral genome, ORF1 and ORF2. Of these, ORF2 codes for structural proteins VP1, VP2, VP3 and VP4, whose masses in the case of Triatoma virus (TrV) are 29.7kDa, 28.4kDa, 32.8kDa and 5.5kDa, respectively1. These are naked viruses, with icosahedral symmetry, approx. 30nm in diameter and positive-sense single-strand RNA with 9010 nucleotides.
TrV, together with the Solenopsis invicta virus (SINV-1), are two members of this family that are pathogenic to medically important insects3. Due to its horizontal mode of transmission and high pathogenicity in triatomine populations, TrV has been proposed as a biological control agent for the vectors of Chagas disease9,10. Recently, this virus has been shown to be not infectious for mice and probably not for other vertebrates either, a finding that reinforces the possibility that it might be used as a tool for the biological control of triatominos12.
Up to the present, the purification protocols used for TrV have been based on the use of whole infected insects, their gut content, their intestines or pieces of paper with dry feces1,7,11. These techniques are time-consuming and not always provide good/high efficiency of recovery; thus, with the goal of optimizing the viral purification technique, this work proposes certain methodological modifications. Obtaining viral mass is of interest in order to enable recovery of viral particles to be used in further trials.
In this work we present a method for viral purification that allows to reduce the necessary time, using a minimal sample. This technique may be applied for TrV and might also be used for other members of the family Dicistroviridae.
Viral particles were purified from fecal matter obtained by abdominal compression of one, five and ten nymphs V from an experimental colony of Triatoma infestans infected with TrV. The insects were fed (10 days after molting into the 3rd stage) during 1h using a solution of human blood and viral proteins of 0.2mg/ml using an artificial feeder and kept at the Centro de Estudios Parasitológicos y de Vectores (CEPAVE), La Plata, Argentina8,9. The samples were identified as follows:
Sample 1 (M1): pooled fecal matter from 10 insects (0.0235g);
Sample 2 (M2): pooled fecal matter from 5 insects (0.0101g);
Sample 3 (M3): fecal matter from 1 insect (0.0035g).
Each sample was individually homogenized with 150μl of phosphate-buffered saline (PBS), stirred for 30s and centrifuged for 10min at 13000×g. The pellet was discarded and the supernatant was again centrifuged for 10min at 13000×g. After this second centrifugation, the supernatants of the samples used (M1, M2) were collected in two tubes for a third and last centrifugation along with M3 for 10min at 13000×g. In this manner, cell remains are discarded and only viral particles are obtained. The supernatant from each sample (M1: 1500μl; M2: 750μl and M3: 150μl) was seeded into three tubes in a continuous sucrose gradient (10–30%, w/v) and centrifuged for 3h at 30000×g at 4°C in a SW65 rotor (Beckman L8-80M Ultracentrifuge). Subsequently, fractions of 500μl from each tube were taken and preserved at −20°C. Twenty-five microliters of each fraction were seeded into 12.5% polyacrylamide gels to corroborate presence of the viral proteins. They were dyed with Coomassie blue and additionally a kit for silver staining (Pierce® Silver Stain Kit).
Later, positive fractions were quantified in 12.5% polyacrylamide gels using an albumin pattern as reference (25μl from each sample).
VP1 and VP2 two of the four structural proteins of TrV were detected by SDS-PAGE, (Fig. 1). According to the albumin pattern of the polyacrylamide gel used as reference, the following concentration results were obtained:
Sample 1 (M1): Approx. 30μg/ml;
Sample 2 (M2): Approx. 20μg/ml;
Sample 3 (M3): Approx. 5μg/ml.
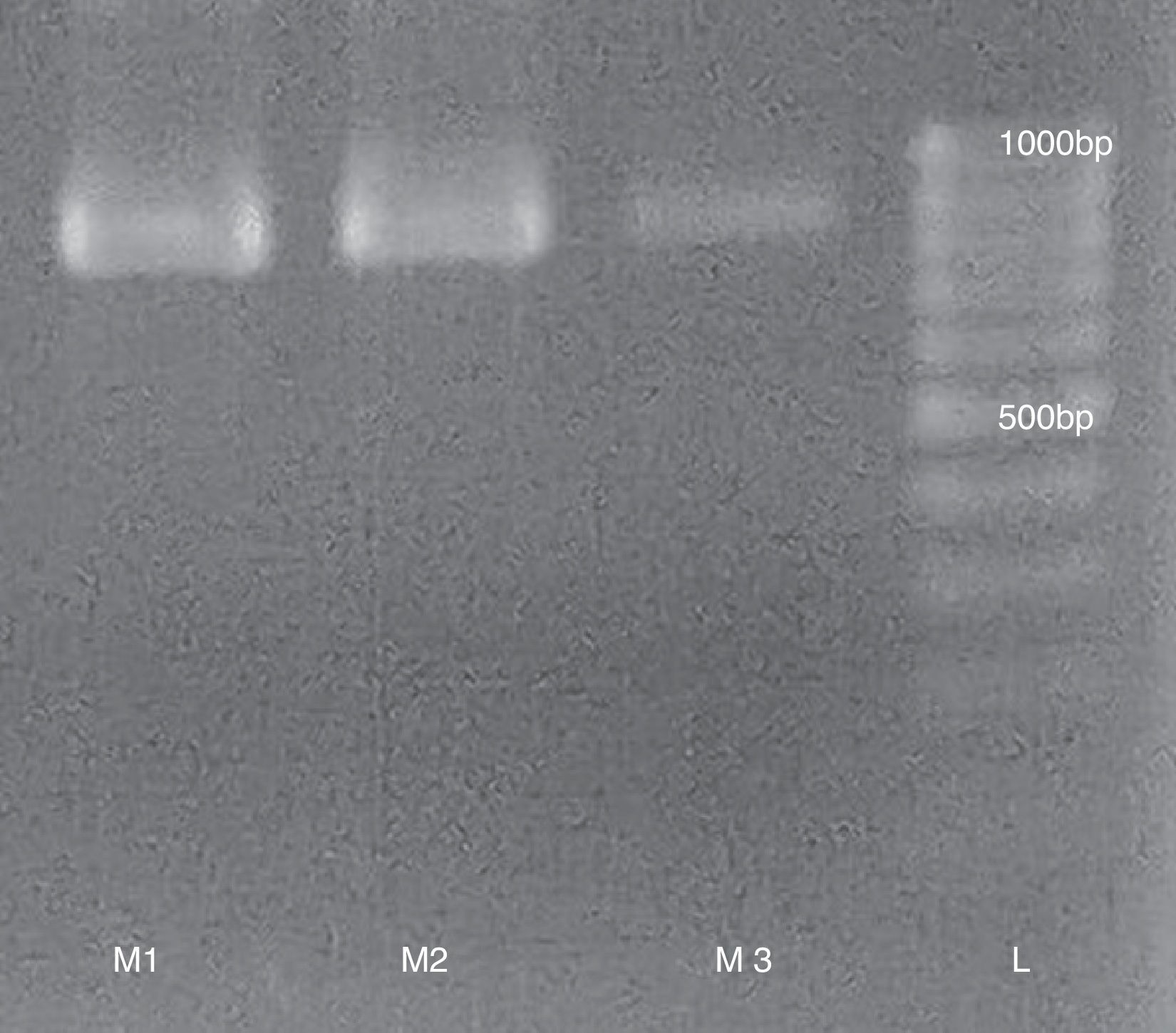
To measure the viral activity from purified samples we performed a RT-PCR to detect the presence of infectious RNA using the following primer pairs: TrVA- 5′ TCAAAACTAACTATCATTCTGG 3′ and TrVB- 5′ TTCAGCCTTATTCCCCCCC 3′ with an expected product of 832bp (Fig. 2). RT-PCR was performed following the methodology proposed by Marti et al7.
The total duration of the procedure described here is 7:30h; in comparison with other techniques for viral purification such as that described by Marti et al.7, which represents a 30% decrease in the total time required, reducing by 50% the time required for the clarification of macerates and dissection of insects. Although the purifications in ClCs used by Muscio et al.11 and Agirre et al.1, are useful to separate RNA-full from empty capsids, they imply adding one more day to the procedure duration.
With regard to procedure duration, obtaining viral particles from fecal matter is similar to using dry fecal buttons1. Starting from fecal matter or buttons allows to decrease contamination during clarification, given that intestinal contents and insect macerate contain lipids and cuticular remains that complicate the procedure. The positive bands obtained in the three samples by RT-PCR indicate infectious viral particles. The speed of our method using fresh fecal matter would allow the viral particles to still remain active and avoid sacrificing insects from the colony.
Up to the present, little is known about this viral family. However, the number of recorded instances of these viruses affecting insect with medical/sanitary and agricultural/economic importance is constantly increasing, and several authors have postulated them as biological control agents2–5,9,10,14.
The development of fast, reliable and inexpensive methods to diagnose TrV infection could be useful for future research applied to other Dicistrovirus. The problems that TrV may cause when introduced into insectaries through triatomine colonies are well known, as is its presence in insectaries in Argentina and Brazil8. This TrV purification procedure is advantageous because it avoids sacrificing the insect colony, currently the only known source of viral particles, and allows direct extraction from very small amounts of fecal matter. This is important because it is still difficult to obtain viral replication in diverse cell culture lines. Although Electron Microscopy (EM) is used for the detection of viral particles13, this is complicate because it requires access to an electron microscope and a good resolution for small particles. Furthermore, the latter technique may have poor sensitivity to low viral loads and requires costly equipment; therefore, it is not suitable for routine analyses.
It is essential to have an adequate (low-cost and fast) method for viral purification, and, compared with traditional methods, the technique proposed here is a good alternative to previously used ones7,11. This method is fast and efficient compared to other proposals, allowing the rapid extraction of a good amount of purified virus from a minimal amount of fecal matter7,11,13.
Ethical disclosuresProtection of human and animal subjectsThe authors state that no human or animal experiments have been performed for this research.
Confidentiality of dataThe authors state that they have followed the protocols of their work center on the publication of patient data.
Right to privacy and informed consentThe authors declare that no patient data appear in this article.
Conflict of interestThe authors declare that they have no conflicts of interest.
This work was partly funded by grants PIP N° 0007 (CONICET) and PICT N° 2014-1536 (ANPCyT). We thank the Virology Department, Facultad de Ciencias Veterinarias, Universidad Nacional de La Plata (UNLP), Argentina, and CEPAVE (Centro de Estudios Parasitológicos y de Vectores -CONICET-UNLP, Argentina) where this study was performed.