Salmonella enterica serovar Heidelberg ranks among the most prevalent causes of human salmonellosis in the United States and Canada, although it has been infrequently reported in South American and European countries. Most Salmonella infections are self-limiting; however, some invasive infections require antimicrobial therapy. In this work we characterized an oxyimino-cephalosporin resistant S. Heidelberg isolate recovered from an inpatient in a Buenos Aires hospital. CMY-2 was responsible for the β-lactam resistance profile. S. Heidelberg contained a 97kb plasmid belonging to the Inc N group harboring blaCMY-2. ISEcp1 was located upstream blaCMY-2 driving its expression and mobilization. The isolate belonged to sequence type 15 and virotyping revealed the presence of sopE gene. In this study we identified the first CMY-2 producing isolate of S. Heidelberg in Argentina and even in South America.
Salmonella enterica serovar Heidelberg es uno de los principales agentes causantes de salmonelosis en humanos en Estados Unidos y Canadá, sin embargo, resulta infrecuente en los países de Sudamérica y Europa. En este trabajo se caracterizó un aislamiento de S. Heidelberg resistente a oximino-cefalosporinas recuperado de un paciente internado en un hospital de la Ciudad de Buenos Aires. Se evidenció la presencia de un plásmido de 97kb perteneciente al grupo de incompatibilidad IncN, portador del gen blaCMY-2. ISEcp1 fue localizado corriente arriba de blaCMY-2, promoviendo su expresión y movilización. El aislamiento de S. Heidelberg correspondió al secuenciotipo 15 y en la virotipificación se detectó el gen sopE. En este trabajo describimos por primera vez la producción de CMY-2 en una cepa de S. Heidelberg en nuestro país y América Latina.
Salmonella enterica serovar Heidelberg is the causative agent of salmonellosis, a self-limiting gastroenteritis that does not usually require antibiotic therapy. However, severe infections may occur, particularly in children and immunocompromised hosts, leading to invasive diseases that require antimicrobial treatment. Fluoroquinolones and extended-spectrum cephalosporins are frequently used in severe Salmonella infections10.
Since the late ‘80s Salmonella isolates displaying resistance to extended spectrum cephalosporins have emerged worldwide. Coding genes for TEM-, SHV-, PSE-, OXA-, PER-, CTX-M-, CMY-, ACC-, DHA- extended spectrum β-lactamases (ESBL) and also KPC carbapenemases have been reported in S. enterica isolates9,14.
S. enterica serovar Heidelberg ranks among the most prevalent causes of human salmonellosis in the United States and Canada, although it is infrequently reported in South American and European countries1,8,9. During the last decade, extended-spectrum cephalosporin resistance has increased among human and agri-food isolates of this serotype in North American countries. This resistance profile is mainly associated with the spread of blaCMY-2 plasmid encoded AmpC β-lactamase10. S. Heidelberg is also one of the most common Salmonella serovars isolated from poultry and eggs, whose consumption has led to many foodborne infection outbreaks. Infections caused by personto- person transmission or direct contact with infected animals have been rarely reported7.
In Argentina, S. Heidelber g isolates are very infrequent among those submitted to the Centro Nacional de Referencia (Mariana Pichel-Instituto Nacional de Enfermedades Infecciosas-ANLIS “Carlos G. Malbrán”-personal communication).
In this study, we characterized oxyimino-cephalosporin resistance in an S. Heidelberg isolate recovered from a diarrheal stool sample of an HIV adult inpatient, in February 2012, in Buenos Aires. Identification was carried out using conventional culture methods. Serotyping was conducted at the Centro Nacional de Salmonella (CNS) in Montevideo, Uruguay. The CNS, housed in the Departamento de Bacteriología y Virología, Instituto de Higiene, Universidad de la República, has characterized Salmonella isolates of human, animal, food, feed and environmental origin, voluntarily submitted by several private and public laboratories for the last 60 years in Uruguay.
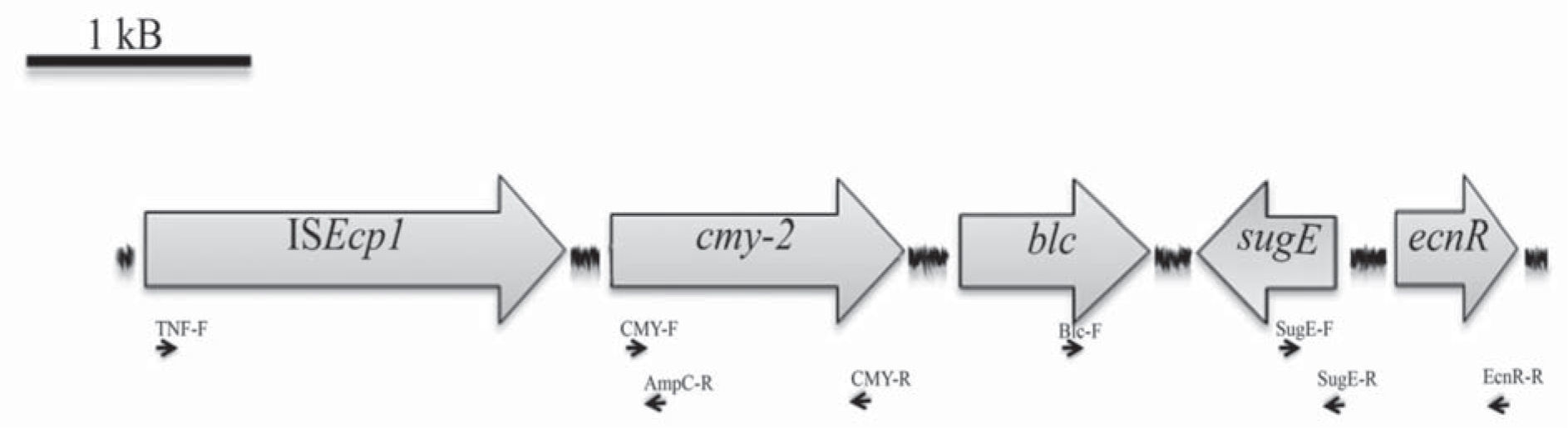
Minimal Inhibitory Concentrations (MICs) of different antimicrobial agents were determined using broth microdilution testing and interpreted according to the Clinical and Laboratory Standards Institute (CLSI) guidelines5. S. Heidelberg was resistant to ampicillin, cephalothin, cefoxitin, ceftriaxone, ceftazidime, intermediate to tetracycline and susceptible to cefepime, imipenem, aztreonam, kanamycin, gentamicin, ciprofloxacin, levofloxacin and cloramphenicol. Phenotypic screening for β-lactamases was performed by synergy tests using amoxicillin/clavulanic acid (10μg/10μg) and phenyl-boronic acid (300μg)-containing disks. Synergy was observed between phenyl-boronic acid and both ceftazidime and cefotaxime disks, suggesting the presence of an AmpC type β-lactamase. Plasmid DNA was purified according to the Kado and Liu method. A multiplex-PCR assay was conducted to reveal the presence of plasmidencoded ampC alleles15, rendering a 462 bp amplicon, which suggested the presence of a coding gene for a CIT cluster β-lactamase. The following specific primers (5′-3′) were used to achieve the complete blaCMY gene: CMY-F: ATGATGAAAAAATCGTTATGCT and CMY-R: TTATTGCAGCTTTTCAAGAATGCG. The nucleotide sequence of the 1140 bp amplicon obtained corresponded to blaCMY-2. The genetic context of blaCMY-2 was determined by PCR mapping and sequencing, as shown in Figure 1, using the following primers (5′-3′): TNF: ACCTAGATTCTACGTCAGTACT, AmpC-R: CCCTGGTAGATAACGGCA, Blc-F: CATTCCTGGTTGTCGCGTGT, SugE-F: AGCATGGCGATACTGACGAT, SugE-R: GCCTGATATGTCCTGGATCGT, EcnR-R: GGATTGAGAGGGCACGAT. ISEcp1 was located upstream blaCMY-2, and blc, sugE and ecnR were identified downstream (Accession number HG931731). The analyzed blaCMY-2 context agrees completely with the conserved regions reported for Type I, II and III environments described in S. enterica, in which blaCMY-2 gene is associated with the insertion sequence ISEcp1, which could enhance blaCMY-2 expression and mobilization13.
Replicon type of blaCMY-2 harboring plasmid was determined according to Carattoli et al.2, corresponding to the IncN group. Plasmid size was estimated in 97 kb by PFGE analysis of S1 nuclease digested DNA9. Conjugation assays were carried out using E. coli J53 (sodic azid resistant) as recipient strain and Luria Bertani agar plates supplemented with sodium azide (150μg/ml) and ceftadizime (10μg/ml) as selection system. blaCMY-2 plasmid could not be transferred by conjugation in the assayed conditions.
Multilocus sequence typing (MLST) with seven housekeeping genes (aroC, dnaN, hemD, hisD, purE, sucA, thrA) was conducted according to http://mlst.ucc.ie/mlst/dbs/Senterica. The isolate displayed the following allelic profile: 2, 7, 9, 9, 5, 9, 12, which corresponds to ST 15, as well as the majority of the S. Heidelberg isolates deposited in the MLST database. According to the S. enterica MLST database, ST 15 was more often reported in Europe, North America and Asia, however there is only one description in Africa and two in South America http://mlst.ucc.ie/mlst/mlst/dbs/Senterica/.
Based on the ST analysis, in 2012, S. enterica isolates were grouped together in 138 discrete genetically related clusters called eBurstGroups (eBGs). Some eBGs exhibit a unique oneto-one relationship with serovars such as eBG26 and S. Heidelberg (http://mlst.ucc.ie/mlst/mlst/dbs/Senterica/).
Virotyping was performed by PCR amplification of coding genes for proteins secreted by type III secretion systems (avrA, sopE), Salmonella Typhimurium genomic island CS54 (shdA) and phage encoded genes (gogB and sb41); specific primers for invA were included as an internal control6. Among the virulence-related genes investigated by PCR amplification, only sopE was detected. The sopE gene encodes for a Rho- GTPase that induces membrane ruffling and elicits a proinfl ammatory response in epithelial cells. The cytosolic localization of SopE in the absence of other bacterial molecules is sufficient for inducing NF-κB activation11.
Although there is a national network of laboratories that conducts an exhaustive surveillance of diarrheal episodes, reports of Salmonella spp. infections are not mandatory, except for S. Typhi. It is estimated that only 5% of salmonellosis infections are registered. According to national reports S. Typhimurium and S. Enteriditis constitute the most prevalent serotypes, being S. Heiderberg only sporadically reported. There are no reported data about extended-spectrum cephalosporin resistance among human S. Heidelberg isolates in Argentina. Here we report the first CMY-2-producing S. Heiderberg human isolate in our country, an even in South America.
blaCMY-2 gene, constitutes the most common marker among extended-spectrum cephalosporin-resistant Salmonella in the United States, mainly mediated by the spread of IncI1 blaCMY-2 plasmid1. This replicon type plasmid has also been described in blaCMY-2 producing S. Typhimurium isolated from children with diarrhea in Uruguay6. More recently IncA/C plasmids have been associated with blaCMY-2 bovine isolates of S. Heidelberg10. However, in the studied isolate blaCMY-2 was located in an IncN plasmid, this replicon type has not been previously associated to blaCMY-2 in Salmonella spp. Even in previous studies performed in E. coli in Argentina, where we reported the association of blaCMY-2 with IncA/C, IncI1, IncFIA/FI, IncK, IncF, IncY and IncBO plasmids, the IncN group was not detected3,4,6.
Considering the wide diversity of Inc/blaCMY-2 associations, the spread of blaCMY-2 may be related to the presence of a transposable element responsible for its mobilization. Additionally, the co-mobilization of blaCMY-2 and sugE increases the possibility of co-selection processes. SugE is a member of the small multidrug resistance (SMR) transporter family, responsible for conferring resistance to antiseptics such as quaternary ammonium compounds and SDS12.
The spread of resistance markers among S. Heidelberg isolates constitutes a risk for the management of severe salmonellosis in clinical practice. Therefore, a better understanding of the pathogen distribution and its antimicrobial resistance is important for the development of strategies to limit salmonellosis due to multidrugresistant strains.
Ethical responsibilitiesProtection of human and animal subjectsThe authors declare that no experiments were performed on humans or animals for this investigation.
Confidentiality of dataThe authors declare that no patient data appears in this article.
Right to privacy and informed consentThe authors declare that no patient data appears in this article.
This work was partially supported by Grants from UBACyT and ANPCyT to M. Radice and G. Gutkind. G. Gutkind and M. Radice are members of Carrera del Investigador Científico (CONICET). D. Cejas was recipient of a doctoral fellowship from CONICET and is now recipient of a postdoctoral fellowship from Fundación Bunge y Born.
Conflicts of interest
The authors declare that they have no conflicts of interest.