The aim of this study was to determine the antimicrobial resistance profiles of indicator bacteria isolated from domestic animal feces. Minimal inhibitory concentration (MIC) was determined by agar dilution. Interpretative criteria on the basis of wild-type MIC distributions and epidemiological cutoff values (ECOFF or ECV) were used according to the ‘European Committee on Antimicrobial Susceptibility Testing’ (EUCAST) data. Results from 237 isolates of Escherichia coli showed reduced susceptibility for ampicillin, streptomycin and tetracycline, the antimicrobials commonly used in intensive breeding of pigs and hens. Regarding all the species of the genus Enterococcus spp., there are only ECOFF or ECV for vancomycin. Of the 173 Enterococcus spp. isolated, only one showed reduced susceptibility to vancomycin and was classified as ‘non-wild-type’ (NWT) population. This is the first report in Argentina showing data of epidemiological cutoff values in animal bacteria.
El objetivo de este estudio fue determinar los patrones de resistencia antimicrobiana en bacterias indicadoras aisladas de muestras fecales de animales domésticos. La concentración inhibitoria mínima (CIM) fue determinada por el método de dilución en agar. El criterio de interpretación usado se basó en la distribución de la CIM y el punto de corte epidemiológico (ECOFF o ECV) de acuerdo con los datos del European Committee on Antimicrobial Susceptibility Testing (EUCAST). Los resultados obtenidos de 237 aislamientos de Escherichia coli mostraron sensibilidad reducida a ampicilina, estreptomicina y tetraciclina, antimicrobianos comúnmente usados en porcinos y aves de explotación intensiva. Con respecto a todas las especies del género Enterococcus spp., solo existe ECOFF o ECV para la vancomicina. De los 173 Enterococcus spp. aislados, sólo uno presentó sensibilidad reducida a dicho agente y fue categorizado como población ‘non-wild-type’ (NWT). Este es el primer informe en Argentina que presenta datos de puntos de corte epidemiológico en bacterias animales.
Antimicrobial resistance is a global public and animal health concern that is influenced by both human and non-human antimicrobial usage. The human, animal and plant sectors have a shared responsibility to prevent or minimise antimicrobial resistance selection pressures on both human and non-human pathogens10. Epidemiologists need to be aware of emerging changes in bacterial susceptibility, which may indicate emerging resistance, and allow for appropriate control measures to be considered3. In an attempt to overcome the problems of differences in interpretative criteria based on clinical or epidemiological data, the European Committee on Antimicrobial Susceptibility Testing (EUCAST) has decided to define separate dividing points for the detection of bacteria with resistance mechanisms and the monitoring of resistance development using wild-type cutoff values (WCV) or epidemiological cutoff values (ECOFF or ECV) and the guidance of therapy via clinical breakpoints9. ECOFF or ECV are determined on the basis of the distribution of minimal inhibitory concentrations (MICs) for an antimicrobial agent and a given bacterial species. EUCAST is in the process of collecting full range MIC data from as many sources as possible. Data are inserted into a database and each distribution is screened for acceptance, and then made freely available5. Although the use of ECOFF or ECV is important for the early detection of decreased susceptibility, this value is inappropriate to determine the percentage of clinical resistance14. This is because there are instances when a bacterial isolate will have a MIC value above the ECOFF or ECV but below the clinical susceptible breakpoint; in this case, such isolate will be clinically susceptible and should therefore not be categorized as resistant but as having decreased susceptibility4. In fact, when reporting data using ECOFF or ECV, the terms ‘susceptible’ or ‘resistant’ are inappropriate; instead, bacteria should be reported as ‘wild-type’ if the MIC or zone diameter falls below the epidemiological cutoff value and as ‘non-wild-type’ if the MIC is higher or the zone diameter is smaller than the epidemiological cutoff value13. Thus, the populations of microorganisms without an acquired phenotypically detectable resistance mechanism are defined as wild-type bacteria and the populations that clearly depart from the ‘wild-type’ populations are classified as ‘non-wild-type’ (NWT).
In Argentina, there is no current antimicrobial resistance surveillance system of public health importance in animals to describe the level of resistance to animal bacteria. In a previous study with the same bacterial isolates used in this work, we determined the antimicrobial susceptibility by the agar diffusion method and showed the results as resistance percentage12.
The purposes of the present study were to determine the antimicrobial resistance profiles of indicator bacteria of animal origin using the wild-type MIC distributions and ECOFF or ECV according to EUCAST data and to begin collecting data that could be used in a future monitoring program.
Escherichia coli and Enterococcus spp. were chosen and collected as indicator microorganisms for susceptibility testing. These bacteria are common commensals, which are considered to constitute a reservoir of antimicrobial resistance genes, which may be transferred to pathogenic bacteria causing disease in animals or humans11. The isolates included in this study were collected from 2006 to 2007. A number of 237 Escherichia coli and 173 Enterococcus spp. were isolated from fifty fecal samples from healthy animals (cattle, horses, sheep, pigs, layer hens and dogs) without clinical signs. Samples were inoculated onto selec tive and differential media according to the bacterial genus11. The Enterococcus species were identified on the basis of yellow pigment production, motility, deamination of arginine, utilization of pyruvate and carbohydrate fermentation of arabinose, sorbose, ribose, raffinose, sucrose and mannitol6. The agar dilution susceptibility test was used to determine the minimal inhibitory concentration (MIC)2 of different antimicrobials. Interpretative criteria were used on the basis of wild-type cutoff values also called “epidemiological cutoff values” (ECOFF or ECV) according to EUCAST data5.
Escherichia coli ATCC 25922, Enterococcus faecalis ATCC 29212 and Staphylococcus aureus ATCC 29213 were used as quality control. For E. coli, the following antimicrobial agents were tested according to the following ECOFF or ECV: ampicillin ≤ 8μg/ml, cephalothin ≤ 32μg/ml, gentamicin ≤ 2μg/ml, amikacin ≤ 8μg/ml, streptomycin ≤ 16μg/ml, nalidixic acid ≤ 16μg/ml, enrofloxacin ≤ 0.12μg/ml, ciprofloxacin ≤ 0.06 μg/ml, chloramphenicol ≤ 16μg/ml, florfenicol ≤ 16μg/ml, tetracycline ≤ 8μg/ml and trimethoprim-sulfamethoxazole ≤ 1μg/ml. For Enterococcus spp., ampicillin, vancomycin, tetracycline, erythromycin and gentamicin were tested.
According to EUCAST, there are no data of ECOFF or ECV in all Enterococcus species for some of the antimicrobials analysed. For E. faecalis and E. faecium, the ECOFF or ECV of ampicillin, vancomycin, tetracycline and erythromycin is ≤ 4μg/ml whereas that of gentamicin is ≤ 32μg/ml. For E. hirae, the ECOFF or ECV of tetracycline is ≤ 4μg/ml whereas that of erythromycin is ≤ 2μg/ml. For E. avium, E. casseliflavus and E. gallinarum, the ECOFF or ECV of ampicillin is ≤ 4μg/ml, whereas for Enterococcus spp. the ECOFF or ECV of vancomycin is ≤ 4μg/ml.
Table 1 shows the distribution of MICs and wild-type cutoff values according to EUCAST for E. coli.Table 2 shows the Enterococcus species and number of isolates among pigs, cattle, sheep, layer hens, horses and dogs. Table 3 shows the MIC distribution for Enterococcus spp.
MIC distribution for Escherichia coli from pigs (n=43), cattle (n=43), sheep (n=18), hens (n=49), horses (n=35) and dogs (n=49).
| ATM | Sample | ≤0.004 | 0.008 | 0.015 | 0.03 | 0.06 | 0.12 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | >128 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | Pigs | 11 | 1 | 7 | 1 | 23 | |||||||||||
| Cattle | 6 | 23 | 13 | 1 | |||||||||||||
| Sheep | 14 | 4 | |||||||||||||||
| Hens | 10 | 10 | 18 | 4 | 1 | 6 | |||||||||||
| Horses | 7 | 20 | 7 | 1 | |||||||||||||
| Dogs | 4 | 10 | 25 | 3 | 1 | 6 | |||||||||||
| CEP | Pigs | 27 | 13 | 3 | |||||||||||||
| Cattle | 1 | 1 | 23 | 18 | |||||||||||||
| Sheep | 5 | 10 | 2 | 1 | |||||||||||||
| Hens | 16 | 29 | 4 | ||||||||||||||
| Horses | 11 | 15 | 8 | ||||||||||||||
| Dogs | 20 | 27 | 2 | ||||||||||||||
| GEN | Pigs | 7 | 22 | 10 | 1 | 1 | 1 | 1 | |||||||||
| Cattle | 21 | 10 | |||||||||||||||
| Sheep | 9 | 7 | |||||||||||||||
| Hens | 22 | 8 | |||||||||||||||
| Horses | 1 | 15 | 8 | ||||||||||||||
| Dogs | 1 | 20 | 18 | 5 | 2 | 1 | 1 | 1 | |||||||||
| AMK | Pigs | 2 | 41 | ||||||||||||||
| Cattle | 4 | 19 | 20 | ||||||||||||||
| Sheep | 1 | 16 | 1 | ||||||||||||||
| Hens | 3 | 31 | 12 | 1 | 2 | ||||||||||||
| Horses | 2 | 19 | 14 | ||||||||||||||
| Dogs | 1 | 24 | 24 | ||||||||||||||
| STR | Pigs | 9 | 7 | 2 | 1 | 6 | 6 | 12 | |||||||||
| Cattle | 19 | 21 | 2 | 1 | |||||||||||||
| Sheep | 1 | 8 | 7 | 2 | |||||||||||||
| Hens | 22 | 17 | 2 | 3 | 5 | ||||||||||||
| Horses | 12 | 18 | 3 | 1 | 1 | ||||||||||||
| Dogs | 21 | 19 | 1 | 1 | 4 | 3 | |||||||||||
| NAL | Pigs | 5 | 10 | 13 | 3 | 3 | 4 | 5 | |||||||||
| Cattle | 1 | 24 | 14 | 4 | |||||||||||||
| Sheep | 17 | 1 | |||||||||||||||
| Hens | 4 | 2 | 13 | 6 | 4 | 20 | |||||||||||
| Horses | 4 | 16 | 12 | 3 | |||||||||||||
| Dogs | 3 | 29 | 12 | 1 | 4 | ||||||||||||
| ENR | Pigs | 12 | 10 | 11 | 3 | 3 | 2 | 1 | 1 | ||||||||
| Cattle | 9 | 920 | 5 | ||||||||||||||
| Sheep | 1 | 5 | 12 | ||||||||||||||
| Hens | 2 | 9 | 1 | 3 | 4 | 10 | 11 | 7 | 1 | 1 | |||||||
| Horses | 6 | 13 | 10 | 6 | |||||||||||||
| Dogs | 19 | 21 | 5 | 1 | 1 | 1 | 1 | ||||||||||
| CIP | Pigs | 18 | 4 | 15 | 1 | 3 | 1 | 1 | |||||||||
| Cattle | 12 | 10 | 16 | 4 | 1 | ||||||||||||
| Sheep | 1 | 16 | 1 | ||||||||||||||
| Hens | 9 | 2 | 1 | 2 | 12 | 4 | 15 | 2 | 2 | ||||||||
| Horses | 7 | 8 | 17 | 2 | 1 | ||||||||||||
| Dogs | 35 | 6 | 4 | 1 | 2 | 1 | |||||||||||
| CMP | Pigs | 1 | 9 | 6 | 6 | 1 | 4 | 2 | 14 | ||||||||
| Cattle | 11 | 20 | 11 | 1 | |||||||||||||
| Sheep | 1 | 1 | 11 | 5 | |||||||||||||
| Hens | 1 | 2 | 34 | 10 | 2 | ||||||||||||
| Horses | 3 | 16 | 14 | 1 | 1 | ||||||||||||
| Dogs | 1 | 24 | 21 | 2 | 1 | ||||||||||||
| FLO | Pigs | 1 | 12 | 11 | 1 | 18 | |||||||||||
| Cattle | 10 | 12 | 21 | ||||||||||||||
| Sheep | 9 | 9 | |||||||||||||||
| Hens | 1 | 28 | 16 | 3 | 1 | ||||||||||||
| Horses | 5 | 14 | 15 | 1 | |||||||||||||
| Dogs | 9 | 15 | 25 | ||||||||||||||
| TET | Pigs | 2 | 2 | 1 | 6 | 32 | |||||||||||
| Cattle | 2 | 27 | 11 | 3 | |||||||||||||
| Sheep | 15 | 1 | 2 | ||||||||||||||
| Hens | 16 | 1 | 4 | 28 | |||||||||||||
| Horses | 1 | 22 | 9 | 1 | 1 | 1 | |||||||||||
| Dogs | 3 | 27 | 5 | 3 | 11 | ||||||||||||
| TMS | Pigs | 23 | 2 | 7 | 2 | 2 | 7 | ||||||||||
| Cattle | 42 | 1 | |||||||||||||||
| Sheep | 18 | ||||||||||||||||
| Hens | 45 | 1 | 3 | ||||||||||||||
| Horses | 33 | 2 | |||||||||||||||
| Dogs | 40 | 3 | 2 | 1 | 1 | 2 |
ATM: antimicrobials, AMP: ampicillin, CEP: cephalotin, GEN: gentamicin, AMK: amikacin, STR: streptomycin, NAL: nalidixic acid, ENR;: enrofloxacin, CIP: ciprofloxacin, CMP: chloramphenicol, FLO: florfenicol, TET: tetracycline, TMS: trimethoprimsulphametoxazole.
The vertical lines indicate wild-type cutoff value.The grey zone indicates the number of bacteria with decreased susceptibility above ECOFF or ECV denominated as ‘non-wild-type’.
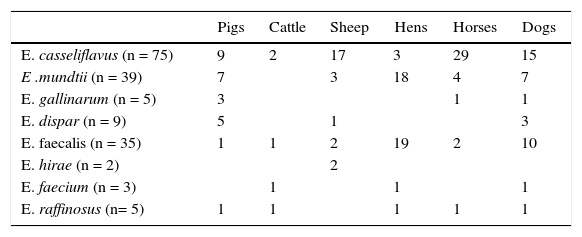
Enterococcus species and number of isolates among pigs, cattle, sheep, hens, horses and dogs.
| Pigs | Cattle | Sheep | Hens | Horses | Dogs | |
|---|---|---|---|---|---|---|
| E. casseliflavus (n = 75) | 9 | 2 | 17 | 3 | 29 | 15 |
| E .mundtii (n = 39) | 7 | 3 | 18 | 4 | 7 | |
| E. gallinarum (n = 5) | 3 | 1 | 1 | |||
| E. dispar (n = 9) | 5 | 1 | 3 | |||
| E. faecalis (n = 35) | 1 | 1 | 2 | 19 | 2 | 10 |
| E. hirae (n = 2) | 2 | |||||
| E. faecium (n = 3) | 1 | 1 | 1 | |||
| E. raffinosus (n= 5) | 1 | 1 | 1 | 1 | 1 |
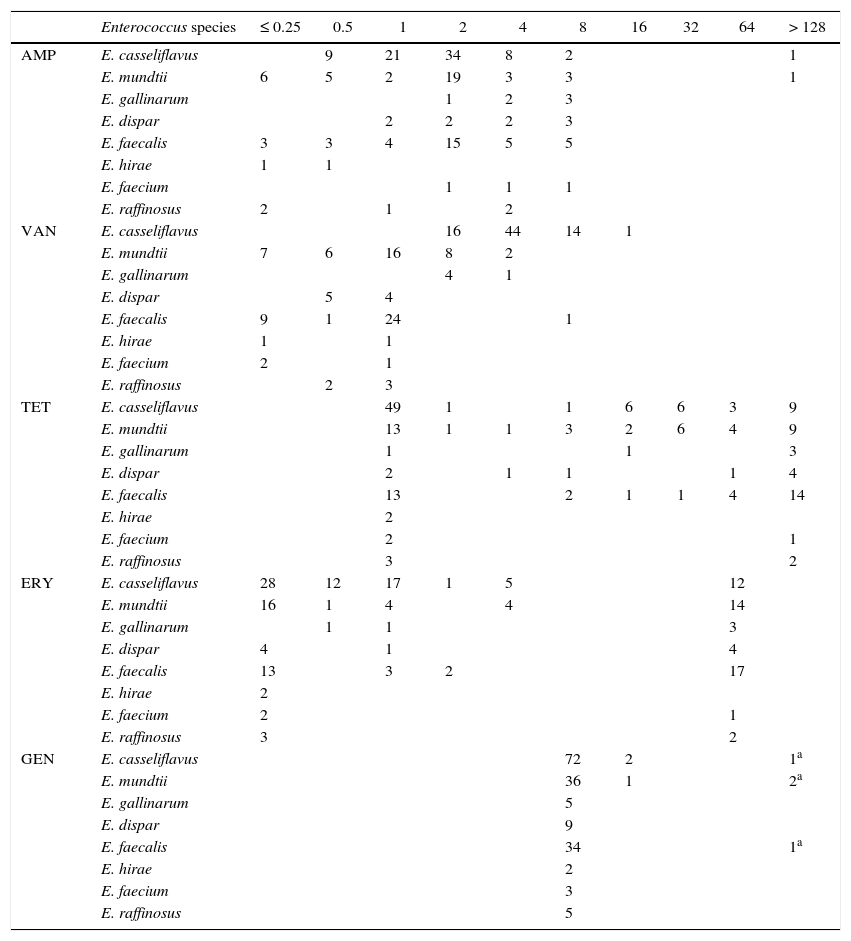
MIC distribution for Enterococcus species from pigs (n=26), cattle (n=5), sheep (n=25), hens (n=43), horses (n=37) and dogs (n=37).
| Enterococcus species | ≤ 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | > 128 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | E. casseliflavus | 9 | 21 | 34 | 8 | 2 | 1 | ||||
| E. mundtii | 6 | 5 | 2 | 19 | 3 | 3 | 1 | ||||
| E. gallinarum | 1 | 2 | 3 | ||||||||
| E. dispar | 2 | 2 | 2 | 3 | |||||||
| E. faecalis | 3 | 3 | 4 | 15 | 5 | 5 | |||||
| E. hirae | 1 | 1 | |||||||||
| E. faecium | 1 | 1 | 1 | ||||||||
| E. raffinosus | 2 | 1 | 2 | ||||||||
| VAN | E. casseliflavus | 16 | 44 | 14 | 1 | ||||||
| E. mundtii | 7 | 6 | 16 | 8 | 2 | ||||||
| E. gallinarum | 4 | 1 | |||||||||
| E. dispar | 5 | 4 | |||||||||
| E. faecalis | 9 | 1 | 24 | 1 | |||||||
| E. hirae | 1 | 1 | |||||||||
| E. faecium | 2 | 1 | |||||||||
| E. raffinosus | 2 | 3 | |||||||||
| TET | E. casseliflavus | 49 | 1 | 1 | 6 | 6 | 3 | 9 | |||
| E. mundtii | 13 | 1 | 1 | 3 | 2 | 6 | 4 | 9 | |||
| E. gallinarum | 1 | 1 | 3 | ||||||||
| E. dispar | 2 | 1 | 1 | 1 | 4 | ||||||
| E. faecalis | 13 | 2 | 1 | 1 | 4 | 14 | |||||
| E. hirae | 2 | ||||||||||
| E. faecium | 2 | 1 | |||||||||
| E. raffinosus | 3 | 2 | |||||||||
| ERY | E. casseliflavus | 28 | 12 | 17 | 1 | 5 | 12 | ||||
| E. mundtii | 16 | 1 | 4 | 4 | 14 | ||||||
| E. gallinarum | 1 | 1 | 3 | ||||||||
| E. dispar | 4 | 1 | 4 | ||||||||
| E. faecalis | 13 | 3 | 2 | 17 | |||||||
| E. hirae | 2 | ||||||||||
| E. faecium | 2 | 1 | |||||||||
| E. raffinosus | 3 | 2 | |||||||||
| GEN | E. casseliflavus | 72 | 2 | 1a | |||||||
| E. mundtii | 36 | 1 | 2a | ||||||||
| E. gallinarum | 5 | ||||||||||
| E. dispar | 9 | ||||||||||
| E. faecalis | 34 | 1a | |||||||||
| E. hirae | 2 | ||||||||||
| E. faecium | 3 | ||||||||||
| E. raffinosus | 5 |
AMP: ampicillin, VAN: vancomycin, TET: tetracycline, ERY: erythromycin, GEN: gentamicin
The vertical lines indicate wild-type cutoff value.
The grey zone indicates the number of bacteria with decreased susceptibility above ECOFF or ECV denominated as ‘non-wild-type’.
Indicator bacteria constitute a natural part of the intestinal flora of many different animal species that easily acquire resistance, and allow to compare levels of antimicrobial resistance among animal populations. In addition, indicator bacteria allow the direct comparison of resistance among different animal species and the analysis of resistance trends over time.
The results obtained showed decreased susceptibility in intensive breeding of pigs and hens. In pigs, 24 NWT of the 43 Escherichia coli strains isolated showed reduced susceptibility to ampicillin and streptomycin, 20 to chloramphenicol, 18 to florfenicol and 38 to tetracycline, coinciding with the antimicrobials most used in pig farms. In hens, 24 NWT of the 49 Escherichia coli strains isolated showed reduced susceptibility to quinolones, 30 to enrofloxacin, 2 to ciprofloxacin and 32 to tetracycline, the drugs most commonly used in this animal species.
In intensive pig and poultry production, animals are kept confined in overcrowded conditions and they are bred and managed for maximum yield. These conditions compromise their health and their immune responses and encourage infectious disease to develop and spread easily7,8. Up to now, without the aid of drugs for disease prevention, it would not be possible to keep the animals productive under these intensive conditions.
In Enterococcus species, there are ECOFF or ECV only for vancomycin (≤ 4μg/ml), and in this study only one E. faecalis isolate was classified as NWT. For tetracycline, a bimodal distribution was observed in some species. Based on the results, 26 E. casseliflavus of the wild-type population were isolated from horses and 8 from dogs, where the selective pressure of antimicrobials is minimal. For E. mundtii and E. faecalis most NWT strains came from layer hens, in which the same behaviour was observed for erythromycin. On the other hand, 75 strains of E. casseliflavus and 5 E. gallinarum were found to have MICs between 2 and 32μg/ml for vancomycin, phenotypically corresponding to the gene VanC, which is characterized by chromosomally mediated, non-transferable, intrinsic low-level resistance to vancomycin1.
Some monitoring programs show the resistance percentage as well as the MIC distribution in their result tables15. In the present study, we decided not to determine this percentage, and, instead, to follow Schwarz's criterion13, which considers that when comparing resistance percentages among published studies, authors must make sure that the same methodologies and the same interpretative criteria have been used.
This study represents the first published data of wildtype MIC distributions of bacteria isolated from different animals in Argentina and we consider that the values herein obtained might serve as a starting point for a future monitoring program.
Ethical responsabilitiesProtection of human and animal subjects. The authors declare that no experiments were performed on humans or animals for this investigation.
Confidentiality of data. The authors declare that no patient data appears in this article.
Right to privacy and informed consent. The authors declare that no patient data appears in this article.
The authors declare that they have no conflicts of interest.