We describe the first isolation of an enteroaggregative Escherichia coli (EAEC) O104:H4 strain associated with an acute diarrhea case in Argentina. Two multiplex PCRs (mPCR) were performed as screening of genes mPCR1 (eae, lt, and st) and mPCR2 (IpaH, aggR, stx1 and stx2). A mPCR to detect the rfbO104, fliCH4 and terD genes, and PCR assays for the detection of pCVD432 plasmid, aaiC and lpfO113 genes were included. Biochemical and antimicrobial susceptibility assays as well as serotyping were performed. The identified E. coli strain was susceptible to all antimicrobials tested and harbored the aggR, aaiC, pCVD432 plasmid, lpfO113, rfbO104, fliCH4 and terD genes. Although serotype EAEC O104:H4 rarely spreads and sporadic cases have been reported, global concern increased after the large-scale outbreak in Europe in 2011. The finding of EAEC O104:H4 reinforces the need for improved methodologies for the detection of all E. coli pathotypes.
Se describe el primer aislamiento de una cepa de Escherichia coli enteroagregativo (EAEC) O104:H4 de un caso de diarrea aguda en Argentina. Se realizaron dos PCR múltiples como tamizaje: mPCR1 para los genes eae, lt y st, y mPCR2 para los genes IpaH, aggR, stx1y stx2. Se incluyó una mPCR para detectar los genes rfbO104, fliCH4 y terD, además de PCR simples para los genes del plásmido pCVD432, aaiC y lpfO113. Se realizaron ensayos bioquímicos, de sensibilidad a los antimicrobianos y de serotipificación. La cepa de E. coli identificada fue sensible a todos los antimicrobianos ensayados y presentó los genes aggR, aaiC, plásmido pCVD432, lpfO113, rfbO104, fliCH4 y terD. Si bien EAEC O104:H4 es un serotipo poco común, se han comunicado casos esporádicos, pero la preocupación global aumentó después del brote masivo ocurrido en Europa en 2011. El hallazgo de EAEC O104:H4 refuerza la necesidad de mejorar las metodologías para la detección de todos los patotipos de E. coli en Argentina.
Diarrheagenic Escherichia coli (DEC) strains are a common cause of diarrhea in humans. DEC strains are classified into five pathotypes: enteropathogenic E. coli (EPEC), enterotoxigenic E. coli (ETEC), enteroinvasive E. coli (EIEC), enteroaggregative E. coli (EAEC), and Shiga toxin-producing E. coli8 (STEC).
EAEC was first described in 1987 during a prospective study of pediatric diarrhea in Santiago, Chile8. In general, EAEC is associated with acute and persistent diarrhea in children and adults from industrialized and developing countries, international travelers and immunocompromised patients14.
The reservoir is unknown, but EAEC strains were isolated from the feces of different animals such as calves, piglets and horses, and contaminated food has been described as a possible vehicle for strain dissemination. Cases associated with EAEC infection are reported to be sporadic, but some outbreaks have been described, although the sources of infection have rarely been identified in those episodes9.
Several EAEC virulence-related genes have been described, however, their role in the clinical outcome of infection has not been completely defined. This E. coli pathotype is characterized by aggregative adherence (AA) to HEp-2 cells in a characteristic “stacked brick” pattern. The AA phenotype is associated with the presence of a 60 MDa plasmid (pAA), and the expression of one out of four distinct aggregative adherence fimbriae9 (AAFI, AAFII, AAFIII, and AAFIV). These AAFs are transcriptionally regulated by an AraC/XylS family activator called AggR. Under the control of aggR is the anti-aggregation protein gene (aap), present on the pAA-plasmid, and encoding a protein called dispersin, which promotes dispersal of EAEC on the intestinal mucosa to establish new foci of infection. AggR is also required to express the Aat dispersin translocator and the chromosomal cluster called aggR-activated island (aai), encoding a type VI secretion system. Other putative virulence factors not regulated by aggR are the EAEC heat-stable toxin 1 (EAST-1, encoded by the astA gene) and a set of toxins, known as serine protease autotransporters of Enterobacteriaceae (SPATEs), which include the cytotoxic plasmid-encoded toxin3,10 (Pet).
The pathogenesis of EAEC infection is not fully understood due to the heterogeneity of the strains. A three-stage model based on in vitro and animal data was proposed. Stage I involves initial adherence to the intestinal mucosa and the mucus layer. Stage II comprises enhanced mucus production, apparently leading to the deposit of a thick mucus-containing biofilm encrusted with EAEC. This may contribute to EAEC diarrheagenicity and, perhaps, to its ability to cause persistent colonization, diarrhea and nutrient malabsorption. Stage III, suggested from histopathologic and molecular evidence, includes the elaboration of toxins or inflammation, which result in damage to the mucosa and intestinal secretion9.
The clinical profile is characterized by watery, mucoid, secretory diarrhea with low-grade fever, with or without vomiting. However, up to one third of patients with diarrhea by EAEC had grossly bloody stools. While malnutrition predisposes to persistent diarrhea, asymptomatic colonization with EAEC might cause malnutrition because of the persistent mucosal damage, becoming a possible and dangerous cycle8.
A few years ago, global concern about EAEC increased when a large-scale outbreak associated with a new Shiga toxin-producing/enteroaggregative E. coli pathotype occurred in Europe in May-June 2011. A total of 881 hemolytic uremic syndrome cases, 32 deaths and 3141 cases of diarrhea were reported. The STEC/EAEC outbreak strain belonged to the O104:H4 sertotype2. Since then, all countries have increased surveillance of this serotype in all diarrhea cases.
The aim of the present study was to describe the first isolation of an EAEC O104:H4 strain associated with a diarrhea case in Argentina.
On January 2013, a 6-year-old girl presented with a 72-hour history of acute diarrhea at Trelew Zonal Hospital, in the Province of Chubut. At the local laboratory, E. coli was isolated from a stool sample, and the strain was then sent to the National Reference Laboratory (NRL) for further characterization.
At the NRL, the confluent growth zone and colony pools were screened by two multiplex PCRs (mPCR). The mPCR1 enabled to characterize eae (EPEC), lt, and st(ETEC), and the mPCR2, IpaH (EIEC), aggR (EAEC), stx1 and stx2 (STEC) genes7. The mPCR for detection of rfbO104, fliCH4, and terD genes was carried out as described2, with minor modifications. By single PCR assays, the sequence of the pCVD432 EAEC plasmid, the putative virulence genes, aaiC, and the long polar fimbria lpfO113, were studied1,3,5. The strain was characterized by biochemical tests6, serotyping11, and antimicrobial susceptibility (nalidixic acid, amikacin, ampicillin, ciprofloxacin, chloramphenicol, streptomycin, gentamicin, tetracycline, trimethoprim-sulfamethoxazole and nitrofurantoin), based on 2011 CLSI guidelines4.
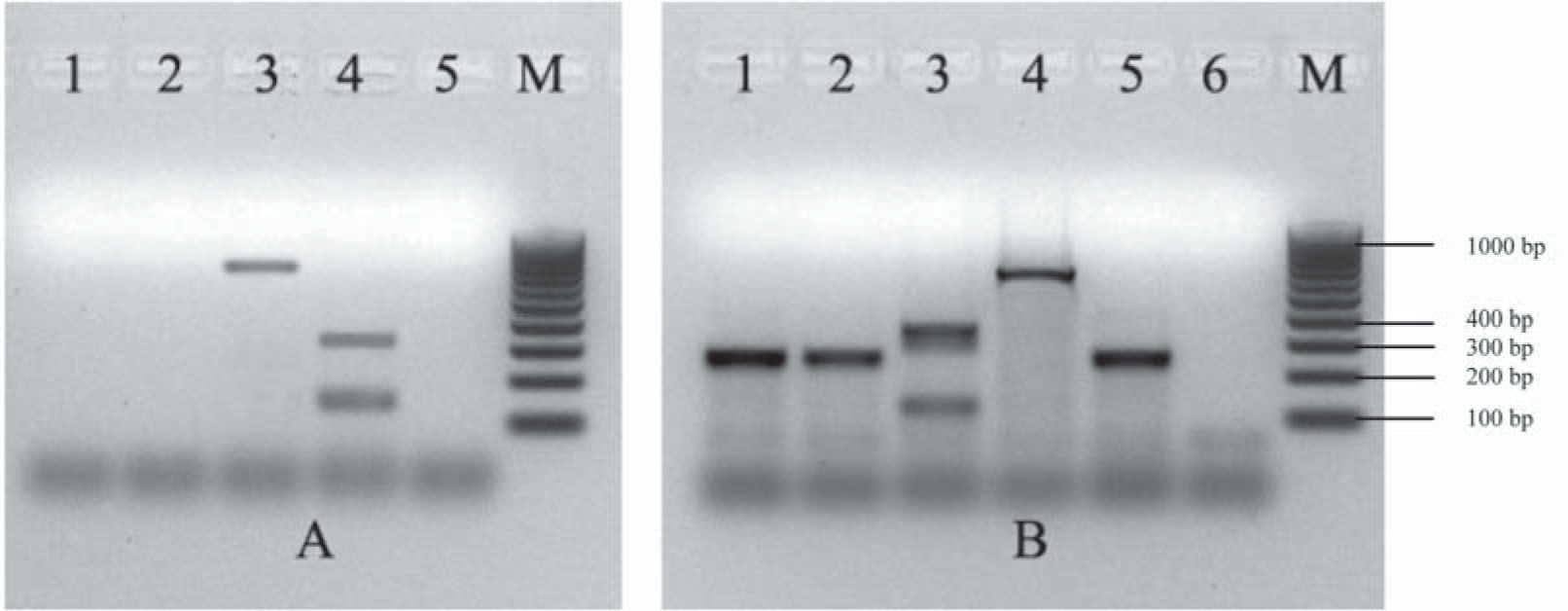
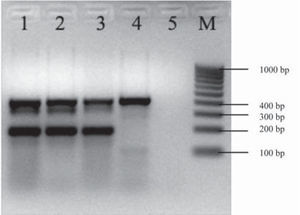
The aggR gene was detected by mPCR1 (Fig. 1). Serotyping and mPCR confirmed that the strain belonged to the O104:H4 serotype and was terD-positive (Fig. 2). The strain harbored the pCVD432 sequence, aaiC and lpfO113 genes. It was identified as E. coli by biochemical tests and was susceptible to all antibiotics tested.
Multiplex PCRs (mPCR) used as screening of EPEC, ETEC, STEC, EIEC and EAEC. A. Multiplex PCR1 for the detection of eae, lt, and st genes. Lane 1: confluent culture of the diarrhea case; lane 2: isolated colony of the diarrhea case; lane 3: E. coli 2348/69 positive control for the eae gene (864 bp); lane 4: E. coli H10407 positive control for lt/st genes (322, 147 bp); line 5: reagent control; M: 100 kb molecular size marker. B. Multiplex PCR2 for the detection of IpaH, aggR, stx1 and stx2 genes. Lane 1: confluent culture of the diarrhea case; lane 2: isolated colony of the diarrhea case; lane 3: E. coli 110/05 positive control for stx1/stx2 genes (130, 346 bp); lane 4: E. coli C-481 positive control for the IpaH gene (619 bp); lane 5: E. coli 17-2 positive control for the aggR gene (254 bp); lane 6: reagent control; M: 100 kb molecular size marker.
Multiplex PCR for detection of rfbO104, fliCH4, and terD genes. Multiplex PCR for the detection of rfbO104, fliCH4, and terD genes. Lane 1: confluent culture of the diarrhea case; lane 2: isolated colony of the diarrhea case; lane 3: E. coli 870 positive control for the terD gene (434 bp), rfbO104 gene (351 bp) and fliCH4 gene (201 bp); E. coli FP595/11 lane 4 positive control for the terD gene (434 bp); lane 5: reagent control; M: 100 kb molecular size marker.
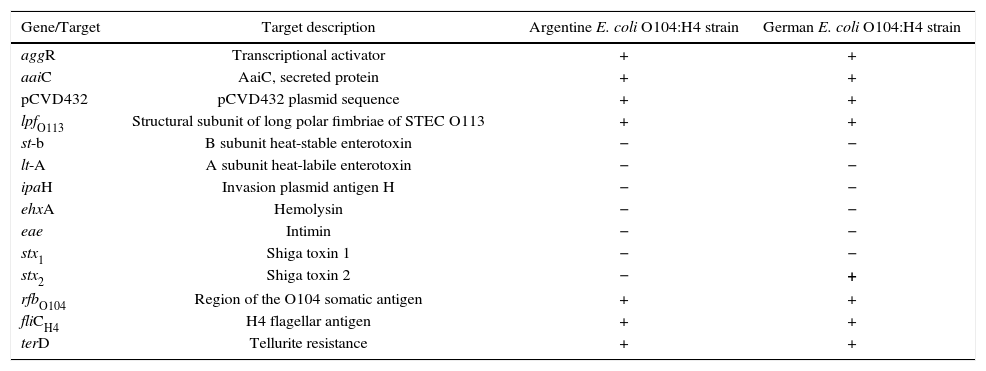
A comparison of the virulence attributes between the Argentine O104:H4 strain and the German outbreak strain is shown in Table 1.
Comparison between the Argentine and German E. coli O104:H4 strains.
| Gene/Target | Target description | Argentine E. coli O104:H4 strain | German E. coli O104:H4 strain |
|---|---|---|---|
| aggR | Transcriptional activator | + | + |
| aaiC | AaiC, secreted protein | + | + |
| pCVD432 | pCVD432 plasmid sequence | + | + |
| lpfO113 | Structural subunit of long polar fimbriae of STEC O113 | + | + |
| st-b | B subunit heat-stable enterotoxin | − | − |
| lt-A | A subunit heat-labile enterotoxin | − | − |
| ipaH | Invasion plasmid antigen H | − | − |
| ehxA | Hemolysin | − | − |
| eae | Intimin | − | − |
| stx1 | Shiga toxin 1 | − | − |
| stx2 | Shiga toxin 2 | − | + |
| rfbO104 | Region of the O104 somatic antigen | + | + |
| fliCH4 | H4 flagellar antigen | + | + |
| terD | Tellurite resistance | + | + |
In the present study, the isolation of an EAEC O104:H4 strain from a diarrhea case was described for the first time in our country.
In Argentina, few data are available on the frequency of EAEC strains in diarrhea cases. In a study performed by Ortiz et al. in Mendoza in 1998, E. coli strains isolated from 140 children under 24 months of age, suffering from acute (n=120), persistent (n=16) and chronic (n=4) diarrhea were studied by the HEp-2 cell-adherence assay. The aggregative adherence (AA) pattern was recognized in strains from 28 (20%) cases. In another study reported by Quiroga et al., the onset time of the first asymptomatic infection caused by the different DEC categories was studied in 44 children since birth until the first 20 months of age in 2000. In all children, at least one DEC strain was detected, and a total of 510 (33.5%) DEC strains were isolated from 1524 fecal samples, with EAEC being the most frequent (31.4%) pathotype identified. Rüttler et al. published the characterization of 87 E. coli strains isolated from patients under 2 years of age, suffering from acute diarrhea in Mendoza in 2002, using the reference HEp-2 assay and AAF/I- and EAST1-PCR assays. Based on the adherence to HEp-2 cells, 22 (25%) strains showed the localized (LA) pattern, 18 (21%) the aggregative (AA) pattern, and 10 (11%) the diffuse (DA) pattern. Twenty-five strains were positive by EAST1-PCR, whereas only 13 of them expressed the AA pattern on HEp-2 cell-adherence assay, and 13 strains were positive by AAF/I-PCR but only 8 showed the AA pattern. More results were reported in 2010 from one study carried out in Corrientes by Medina et al., where 120 children aged 1 month to 14 years with acute diarrhea were cared for in health centers in poor neighborhoods of the city. All children had urban housing with drinking water supply and toilet. DEC strains were detected in 41 (37%) samples, and EAEC strains in 17.1% of the cases12.
The EAEC O104:H4 strain described in the present study harbored the aggR, aaiC, pCVD432 plasmid, lpfO113, rfbO104, fliCH4 and terD genes, as the STEC/EAEC isolated during the outbreak in Europe, however, it was negative for the stx2 gene as is shown in Table 1.
The Argentine strain was susceptible to all the antimicrobial agents tested, while the O104:H4 isolated in Germany showed an ESBL phenotype (resistant to all penicillins and cephalosporins and susceptible to carbapenems), a CTX-M-15 genotype2 and, was also resistant to co-trimoxazole (trimethoprim-sulfamethoxazole) and susceptible to fluoroquinolones (ciprofloxacin) and aminoglycosides (gentamicin, tobramycin).
Serotype EAEC O104:H4 is not commonly spread. Sporadic cases were reported before the 2011 outbreak. Two isolates from patients with HUS in Germany in 2001, one in France in 2004, one from a HUS case in Korea in 2004, two from HUS cases in the Republic of Georgia in 2009, and one from a diarrhea case in Finland in 2010 were reported. The isolates from Germany, Finlandand the Republic of Georgia were EAEC/STEC13. In a study of childhood diarrhea in Mali, EAEC O104:H4 was isolated from three children with moderate to severe diarrhea3.
The finding of an EAEC strain of serotype O104:H4 reinforces the need to implement methodologies for the detection of all E. coli pathotypes. Although O104:H4 is an uncommon serotype, the 2011 European outbreak caused great damage to public health, marking a turning point in terms of surveillance. For this reason, it is important to emphasize that the discovery of new pathotypes is possible, due to the ability of E. coli strains to horizontally acquire specific genetic elements known as pathogenicity islands (PAI), and stxgenes from free bacteriophages in the environment and in mammalian hosts; with an improved adaptation of the bacteria to human hosts. Due to the behavior of this genome, the Reference Laboratories must be able to detect and characterize the emerging new pathogens.
Ethical disclosuresProtection of human and animal subjects. The authors declare that no experiments were performed on humans or animals for this study.
Confidentiality of data. The authors declare that no patient data appear in this article.
Right to privacy and informed consent. The authors declare that no patient data appear in this article.
Conflicts of interestThe authors declare that they have no conflicts of interest.