Microsporidia are obligate intracellular fungi with a remarkable ability to infect a wide range of invertebrate and vertebrate hosts. Namely, Enterocytozoon bieneusi is the most frequently microsporidia reported worldwide, and mainly associated with chronic diarrhea and wasting syndrome in AIDS patients. Microscopy and PCR-based detection techniques are effective for diagnosis and identification of species and genotypes; however, these methods should be standardized in each laboratory. In this study, we performed microscopy and nested PCR techniques with PCR product sequencing to detect E. bieneusi in human stool samples. These techniques, if applied together, might prove useful for diagnosis and future epidemiological studies of intestinal microsporidiosis in Argentina.
Los microsporidios son hongos intracelulares obligados con una notable capacidad para infectar una amplia gama de hospedadores invertebrados y vertebrados. Enterocytozoon bieneusi es el microsporidio más frecuentemente reportado en todo el mundo, principalmente asociado con diarrea crónica y síndrome debilitante en pacientes con sida. Las técnicas de detección basadas en microscopía y PCR son útiles para el diagnóstico y la identificación de especies y genotipos, pero estos métodos deben estar estandarizados en cada laboratorio. En este estudio evaluamos técnicas de microscopía y PCR anidada, con secuenciación de los productos, para detectar E. bieneusi en muestras de heces humanas. Estas técnicas, usadas conjuntamente, podrían ser útiles para su aplicación en el diagnóstico de microsporidiosis intestinal y para realizar estudios epidemiológicos de esta afección en Argentina.
Microsporidia are obligate intracellular parasites with a remarkable ability to exploit a wide range of invertebrate and vertebrate animals, causing from cryptic, benign infections to massive infections with extensive damage leading even to host death5. These organisms were previously considered “primitive” protozoa; however, molecular phylogenetic studies as well as biological evidence have demonstrated that microsporidia are related to the kingdom Fungi, either as a basal branch or sister group5. Although there are 8 genera and 14 species able to infect humans, two species are the most commonly identified, Enterocytozoon bieneusi and Encephalitozoon intestinalis. Human infections are thought to be mainly zoonotic or waterborne and microsporidia spores are released into the environment with feces, urine, and respiratory secretions5. Immunocompetent individuals have asymptomatic or self-limited infections, occasionally reported as “traveler's diarrhea”; however, AIDS patients or other immunosuppressed individuals suffer intestinal microsporidiosis with chronic diarrhea, malabsorption syndrome or severe disseminated disease5. The prevalence of this emerging opportunistic infection in human ranges from 1 to over 50%, depending on the geographic region, the methods applied for diagnosis and demographic characteristics of the studied population11. Albendazole is effective against many species including Encephalitozoon spp., but it shows low efficacy in controlling E. bieneusi disease5.
Transmission electron microscopy (TEM) is regarded as the golden standard for establishing the diagnosis but is expensive, time-consuming and has low sensitivity. Light or immunofluorescence microscopy and molecular amplification techniques, primarily using PCR, are now routinely used for diagnosis. As a matter of fact, PCR assays are significantly more sensitive than light microscopy and provide species differentiation, thus having an impact on treatment decisions3.
In Argentina, 120000 people live with Human immunodeficiency virus (HIV)/acquired immune deficiency síndrome (AIDS) and 6500 new cases are reported annually. In turn, 27.5% of these people are diagnosed late; therefore, they are often at risk of acquiring an opportunistic infection7. Nevertheless, there are few epidemiological data on microsporidiosis13, and a reliable diagnosis is particularly necessary for AIDS patients with chronic diarrhea12. In this study, we standardized microscopy techniques as well as a nested PCR to detect microsporidia in stool samples2,8 so as to be further applied in diagnosis and epidemiological studies in public health in Argentina.
Stool samples (Sample A and B, non-HIV) from Rawson Hospital (Córdoba, Argentina) stock collection samples were used as negative controls and the E. bieneusi positive sample was provided by the Parasitology Laboratory of Hospital Garraham, Buenos Aires, Argentina. The research protocol was approved by the Ethical Committee for Research of Rawson Hospital, Córdoba, Argentina (number 05072018).
Chromotrope 2R technique was performed by following the method described by Weber et al.14, with slight modifications. Briefly, stool samples were fixed with ethanol 70% (1/3 ratio) and then concentrated by the modified ethyl-acetate stool-concentration method4. Smears were fixed with methanol for 5min and incubated overnight with chromotope 2R stain, prepared as described by Weber et al.14 After staining, slides were rinsed and de-colored by dipping in acid alcohol for 10s (moving the slide up and down) and stopped with 95% alcohol. Smears were successively dehydrated in 95% alcohol (5min), 100% alcohol (10min) and xylene (10min). In parallel smears, calcofluor white staining was also performed as previously described6.
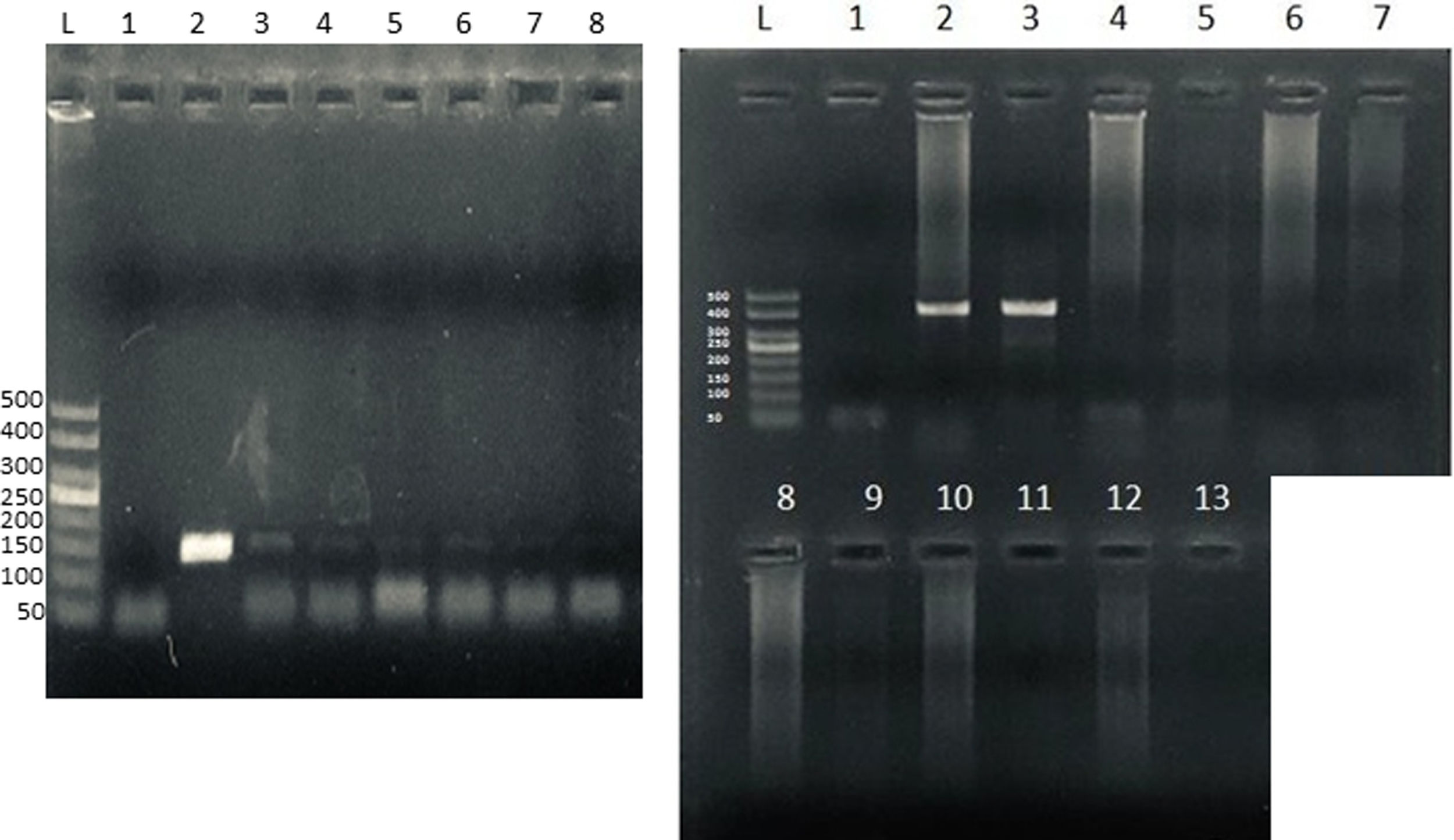
To confirm microsporidia detection by light microscopy, we further performed a nested technique as previously described1 and modified by Santin-Duran et al.2,9,10. DNA extraction was performed by using the DNAeasy Blood & Tissue Kit (Qiagen, USA), according to the manufacturer's instructions. PCR was performed using a set of nested primers specific for E. bieneusi that amplified the ITS and portions of the flanking large and small subunits of the rDNA (∼400bp). The outer primers were EBITS3 (5′-GGTCATAGGGATGAAGAG-3′) and EBITS4 (5′-TTCGAGTTCTTTCGCGCTC-3′), and the inner primers were EBITS1 (5′-GCTCTGAATATCTATGGCT-3′) and EBITS2 (5′-ATCGCCGACGGATCCAAGTG-3′)2. The reaction mixture (20μl) contained 2μl of buffer 10× (16mM MgCl2, 500mM KCl, 100mM Tris–HCl; Dongsheng Biotech, China); 0.16μl of MgCl2 50mM; 0.4μl of 10mM dNTPs; 20pmol of each primer; 0.2μl of 5U/μl of Taq polymerase (Dongsheng Biotech); and 5μl of purified DNA. After denaturation at 94°C for 3min, the PCR samples were subjected to 35 cycles of amplification (denaturation at 94°C for 30s, annealing at 57°C for 30s, and elongation at 72°C for 40s), followed by a final extension at 72°C for 10min. Nested PCR conditions were identical to those of the first run except that only 30 cycles were carried out with an annealing temperature of 55°C. Pure and diluted (1/10, 1/100 and 1/100) PCR products were used in the nested PCR. A negative control without DNA was included in all PCR sets. Furthermore, to rule out PCR inhibition, DNA amplification from Candida albicans was also performed in all samples (Supplementary Fig.). PCR products were subjected to electrophoresis in 2% agarose gel and visualized by staining the gel with ethidium bromide.
To determine E. bieneusi identity, the amplification product from the nested PCR was then subjected to a direct nucleotide sequencing reaction in both directions by using the inner-nested PCR primers (EBITS1 and EBITS2) in the Applied Biosystems® 3500xL Genetic Analyzer at Laboratorio Central de la Provincia de Córdoba, Argentina. Sequences obtained were aligned and inspected by using the MEGA software (version 6), and then compared with reference sequences from the GenBank database by the BLAST analysis10.
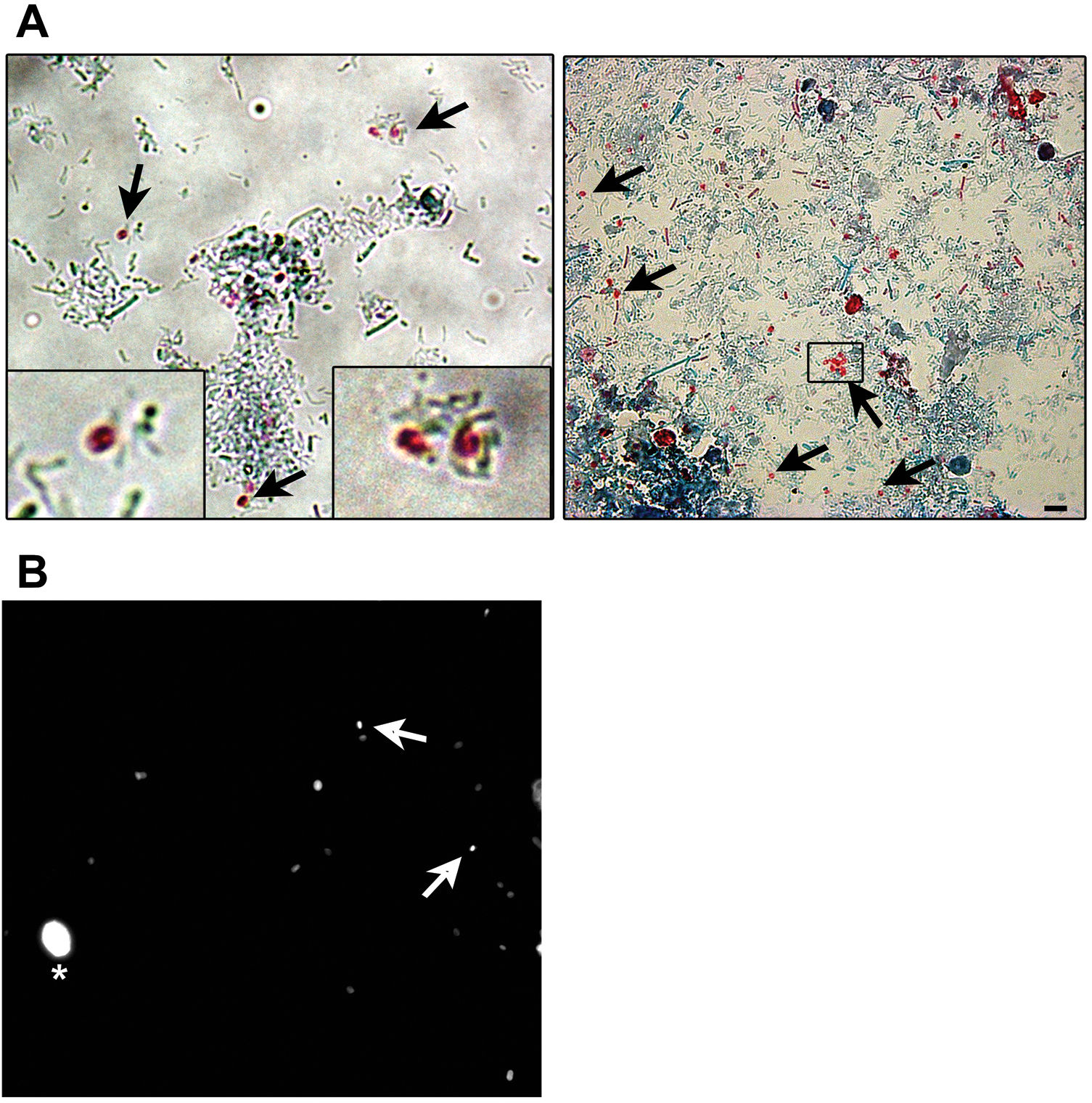
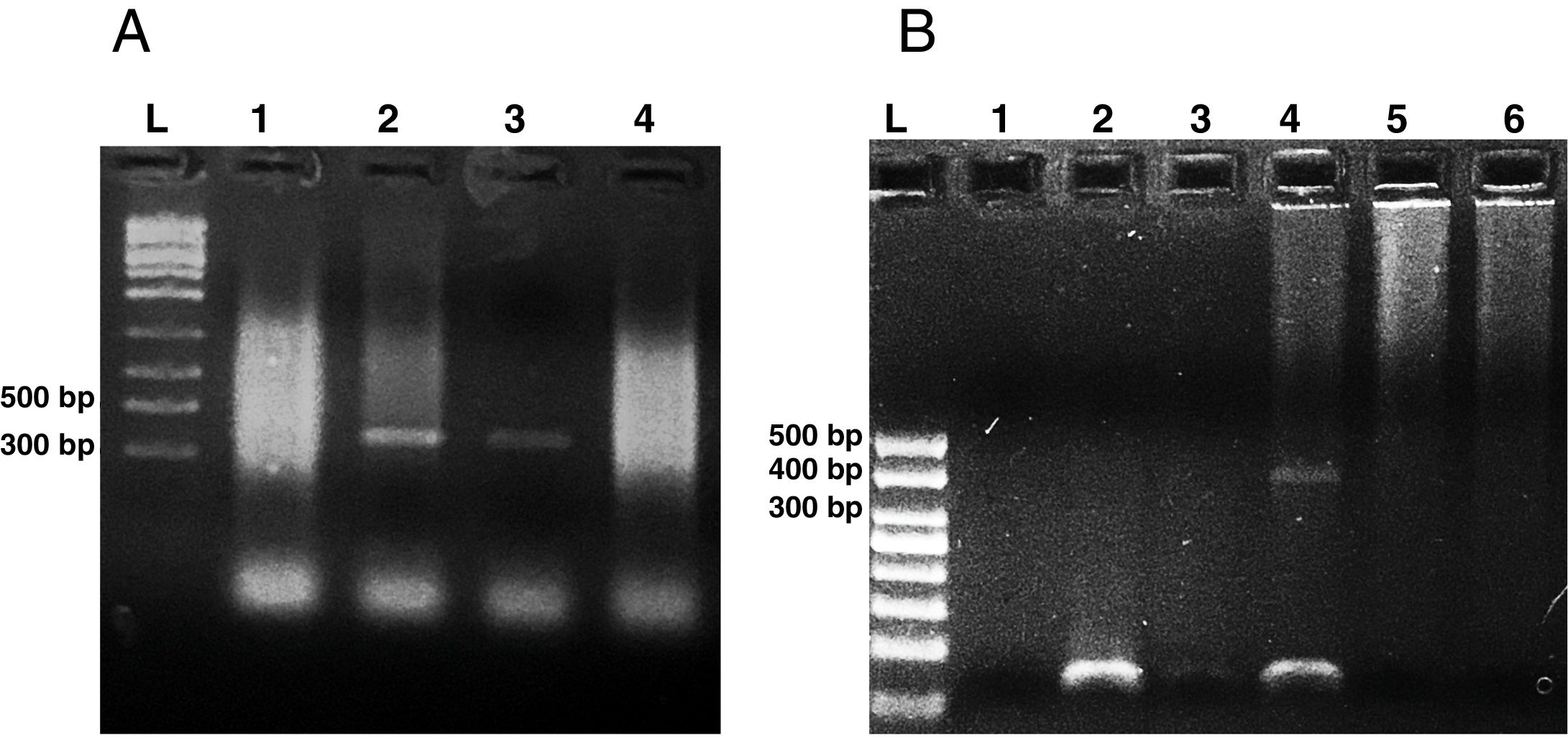
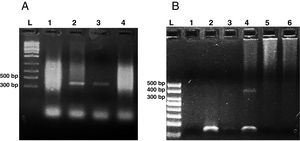
Microsporidia are still difficult to diagnose even though significant progress has been made over the last few years. Nowadays microscopic or PCR-based detection techniques, which must be standardized in each laboratory4, are useful for diagnosis and genotyping. In this study, we standardized the most commonly used staining technique for the examination of stool specimens: chromotrope 2R-based (Weber's modified trichrome) stain14 and chemifluorescent optical brightening agent calcofluor white (that detects chitin in the spore cell wall)4. Figure 1A shows pinkish red-stained microsporidia spores, with some of them showing a distinct pinkish belt-like stripe (1000×). The spores show a characteristic staining pattern and can be readily distinguished from bacteria and fecal debris4,14. In addition, as described by Weber et al.14, calcofluor stained not only microsporidia spores but also yeasts and some feces debris (Figure 1B). In this regard, previous studies comparing the chromotrope staining technique with methods using chemifluorescent optical brighteners have shown that these tests are robust for routine use4. However, none of these microscopic methods are sufficient to determine microsporidia species; therefore, we also performed a nested PCR that is currently used to identify E. bieneusi genotypes and other microsporidia from stool samples. Figure 2 shows that nested PCR produced fragments of approximately 390bp (Fig. 2A and B). Furthermore, the best performance was obtained when products from the first amplification reaction (435bp, not shown) were diluted (1/100 and 1/1000) to be used in the nested PCR (Fig. 2A).
Weber's modified trichome (A and B) and calcofluor (C) staining showing Enterocytozoon bieneusi spores in feces. (A) Modified trichrome staining observed by light microscopy showing ovoid shaped-spores with a pinkish-red stained wall (arrows). Some spores show a pinkish belt-like stripe. (B) Calcofluor staining observed by fluorescence microscopy showing positive E. bieneusi spores (arrows) and a yeast (asterisk). Magnification 1000×. Bars=5μm.
Agarose gel electrophoresis (2%) showing amplification products from the nested from ethanol-fixed stool samples. (A) PCR from Enterocytozoon bieneusi positive stool sample. L: molecular weight ladder. Lanes 1–3: products of nested PCR (with inner primers ITS1 and ITS2) using diluted products from PCR as DNA template (Lane 1: 1/10, Lane 2: 1/100, Lane 3: 1/1000). Lane 4: PCR product using undiluted DNA template from PCR. (B) Nested PCR from different stool samples. L: molecular weight ladder, Lane 1: negative control (H2O), Lane 2: negative stool sample A (PCR product diluted 1/100), Lane 3: negative stool sample B (1/100), Lane 4: positive stool sample (1/100), Lane 5: negative stool sample A (diluted 1/10), negative stool sample B (diluted 1/10).
On the other hand, this PCR technique amplifies the internal transcribed spacer (ITS) region of the rRNA, which has been useful in many studies for the identification of E. bieneusi genotypes2,3,8–10. Accordingly, the sequence analysis of PCR-amplified products showed 99.68% (319 pb) homology with genotype B of E. bieneusi (GenBank accession number AF101198.1)10.
In conclusion, in this work we successfully detected E. bieneusi microsporidia from a human stool sample by using the Chromotrope 2R microscopic technique and a previously reported nested PCR, which can also be applied for E. bieneusi genotyping2,8–10. Further studies are currently underway to define whether these techniques could be useful for human microsporidiosis surveillance in Argentina.
FundingThis work was supported by the Secretary of Scienceand Technology of the National University of Cordoba (Sec-retaría de Ciencia y Tecnología de la Universidad Nacionalde Córdoba, Argentina, SECyT-UNC, PROYECTO CONSOLIDARN∘33620180100408) and the National Agency for Scien-tific and Technological Promotion --- Fund for Scientific andTechnological Research (Agencia Nacional de PromociónCientífica y Tecnológica, Fondo para la Investigación Cien-tífica y Tecnológica, Argentina, FONCyT, PICT 2015-1425).CJM is a fellow from SECyT-UNC (Grants for Socio-productiveTechnological Innovation - Becas de Innovación TecnológicaSocio-Productiva, BITS 2018) and LSC is a member of thescientific career of the National Council for Scientific andTechnological Research (Consejo Nacional de InvestigacionesCientíficas y Tecnológicas, CONICET), Argentina.
Conflict of interestThe authors declare that they have no conflicts of interest.
We wish to thank Dr. Mónica Santin-Duran (United States Department of Agriculture, Maryland, USA) for providing PCR protocols and Dr. Sebastian Dambolena (IMVIB-CONICET, Córdoba, Argentina) for technical support.