In the present study, pentastomids belonging to the order Cephalobaenida were isolated from the lungs of Berber skinks Eumeces schneideri (Famiy: Scincidae), which were morphologically described by light and scanning electron microscopy and taxonomically justified by 18s rDNA molecular analyses of the parasites. Seventeen host specimens were collected from well–vegetated wadis at high altitudes, Jizan, Saudi Arabia as new type locality; twelve specimens (70.59%) were infected. All of the recovered parasites were adults, possessed small broadly triangular cephalothorax flattened on the ventral surface and merged smoothly with a uniformly thick and squat abdomen and terminated in a pair of divergent lobes. The results obtained indicated that the parasites belong to the sharp–tipped posterior–hook Raillietiella spp. distinguished from other raillietiedids of the same group some important characteristic features including annulus number, shape and dimensions of the buccal cadre, copulatory spicules, and anterior and posterior hooks. The anterior hook of the female specimens (n=5) had a blade length (AB) of 135±5 (110–146) μm and shank length (BC) 158±5 (150–169) μm while the posterior hook was much larger with AB measuring 221±5 (200–236) μm and BC 286±6 (280–289) μm. For the male specimens (n=5), the anterior hook had an AB of 73±3 (72–75) μm and a BC 102±5 (100–103) μm. The posterior hook was much larger with AB 190.6±5 (190–191) μm and BC 221±5 (280–289) μm. The morphological characterization of the recovered parasites was closely similar to R. aegypti previously isolated from the same host. Sequence alignment by the maximum likelihood analysis for the data obtained from the 18S rDNA analysis of the parasites exhibits identities ranging between 88–95% with pentastomid genera recovered from the GenBank. The phylogenetic tree supported the inclusion of the parasites within the monophyletic Pentastomida clade with maximum identity to the raillietiellid species. The recovered sequences from the present study were deposited in GenBank under Accession number MK970649.1. The present molecular analysis was the first to confirm the taxonomic position of R. aegypti isolated from the host examined.
En el presente estudio se aisló un pentastómido perteneciente al orden Cephalobaenida del pulmón de un eslizón bereber (Eumeces schneideri, Familia: Scincidae). Se efectuó su descripción morfológica basada en observación por microscopía óptica y de barrido y se justificó su ubicación taxonómica mediante análisis molecular del gen 18S del ADNr. Se recolectaron 17 especímenes del citado huésped en valles ubicados a elevadas altitudes, en la región de Jizan (Arabia Saudí); 12 de ellos (70,59%) estaban infectados. Todos los parásitos recuperados eran adultos y poseían un pequeño cefalotórax triangular, aplanado en la superficie ventral, que se fusionaba con un abdomen abultado y terminado en un par de lóbulos divergentes. Los resultados indicaron que este parásito pertenece a Raillietiella spp., que agrupa especies con gancho posterior puntiagudo; estas se distinguen de otros miembros de la Familia Raillietiella por algunos rasgos característicos, como el número de anillos y la forma y dimensiones del cuadro bucal, las espículas copulatorias y los ganchos anterior y posterior. La caracterización morfológica demostró que el parásito recuperado era muy similar a R. aegypti, previamente aislada del mismo huésped. El alineamiento de secuencias mediante el método de probabilidad máxima basado en el análisis del gen 18s del ADNr detectó identidades del 88–95% con los géneros de pentastómidos disponibles en GenBank. Dentro del árbol filogenético se pudo incluir este parásito dentro del clado monofilético pentastómido con máxima identidad con las especies de Raillietiella. Las secuencias obtenidas fueron depositadas en GenBank, con número de acceso MK970649.1. El presente análisis molecular confirma por primera vez la posición taxonómica de Raillietiella aegypti, anteriormente aislado del mismo huésped.
Pentastomida17 is a subclass of wormlike crustacean endoparasites with five anterior appendages one mouth, and two pairs of hooks for their attachment to the host. They have approximately 131 species distributed in two orders; Cephalobaenida and Porocephalida and seven families: Cephalobaenidae, Linguatulidae, Porocephalidae, Rallietiellidae, Reighardiidae, Sebekidae and Subtriquetridae2, in addition to three Middle Cambrian fossil groups9. The two orders can be distinguished mainly by differences in the disposition of the hooks relative to the mouth and by the form of the reproductive tract1. All of the species of these groups attain sexual maturity in the respiratory tract of vertebrates.
Parasites of the order Cephalobaenida are characterized by the presence of a subterminal or ventral mouth situated anterior to the hooks disposed in a trapezium5,14. Three genera of the order Cephalobaenida were recorded: (a) Cephalobaena34, infecting the lung of American tree snakes; (b) Reighardia Ward, 1899 infecting the lung of marine birds as definitive hosts12,20; (c) Raillietiella53, the largest pentastome, worm–like, blood sucking parasites inhabiting the upper respiratory tract of insectivorous small lizards29,40,46, which have piercing mouthparts surrounded by two pairs of compound hooks that embed into the lung to facilitate feeding on the host blood36,51.
The life cycle of pentastomids includes two hosts; the eggs are either coughed out by the host or leave the host body through the digestive system. The eggs are then ingested by an intermediate host, which is commonly either a fish or a small herbivorous mammal. The definitive host becomes infected by pentastomids after eating the intermediate host, the parasite crawls within the host body into the respiratory tract from the esophagus2,37,52. Humans can serve as an accidental definitive host after ingesting raw or poorly cooked viscera (i.e., liver, lungs, and trachea) of the intermediate hosts22.
The pathologic effect of pentastomid infection may be very serious and/or lethal, causing lesions in lung tissues and obstruction of the trachea that can result in interstitial subacute pneumonia with congestion36,38,49. In Egypt, rallietiellids were first recorded by Ali et al. 3, who described two pentastomids, R. affinis54 and R. aegypti as a new species from Egyptian desert lizards, with much focus on R. affinis. Scant information with incomplete morphology and phylogeny is available worldwide for R. aegypti. The nuclear18S ribosomal DNA (18S rDNA) has proved to be a potential marker during the characterization of eukaryotes23,41. Its highly conserved nature and slow rate of evolutionary mutation make 18S rDNA suitable for interspecies distinction at higher taxonomic levels57.
In the present study, a morphological description of a pentastomid infecting the lungs of Berber skinks Eumeces schneideri (Scincidae) captured in Jizan, Saudi Arabia was carried out by light and scanning electron microscopy. Furthermore, the 18S rDNA molecular analysis of the parasite was conducted to determine the exact phylogenetic position of these parasites.
Materials and methodsSample collection and parasitological studySeventeen specimens of the berber skink Eumeces schneideri (Reptilia: Scincidae), 10 males and 7 females, were collected by hand or noose from well–vegetated wadis at high altitudes, in Jizan, Saudi Arabia (17.6548° N, 42.8871°E) during the year 2017. Specimens were collected seasonally, during summer (5 males, 4 females) and winter (5 males, 3 females). Animals were kept alive in glass cages with sand and alluvium in the animal room at 25–30°C and fed insect larvae. Identification of the examined species was carried out according to Arnold11and Al–Sadoon10. When examined, a cotton ball soaked with isoflurane was placed inside a plastic bag containing the animals and left inside to allow sufficient time for the anaesthetic gas to cause euthanasia. Alternatively, the animals were left in the sealed bag for sufficient time to achieve anaesthesia, at which time they were removed from the bag and injected intraperitoneally with an overdose of sodium pentobarbital. These were in agreement with the regulatory laws regarding experimental ethics of animal use and collecting permits, Institute of Animal Care and Use Committee (protocol number BSU/FS/2015/10).
Following dissection, pentastomids were recovered from lungs using a ZEISS Compact Greenough stereomicroscope (Model Stemi 305), heat fixed in 10% neutral buffered formalin for 15min and then preserved in 70% ethanol in 5% glycerol solution to avoid sudden drying. Finally, samples were transferred to lactophenol for clearance. The prepared samples were examined using differential interference contrast (DIC) light microscopy with digital image analysis system (analysis auto 5.0) and photographed by an Olympus research photomicroscope supplied with a built–in camera (Model BX53F, Tokyo, Japan). Drawings were made with the aid of a drawing tube.
For scanning electron microscopy, samples were fixed in 3% glutaraldehyde in 0.1M sodium cacodylate buffer, washed in the same buffer, and dehydrated in a graded alcohol series. Samples were then processed in a critical point drier “Bomer–900” with freon 13, sputter–coated with gold–palladium in a Technics Hummer V, and finally examined with a Jeol scanning electron microscope (Model JSM7610F, Jeol Ltd, Japan). Identification of the recovered pentastomids was based on the key published by Rego48and Ali et al.4,6.
Molecular analysesGenomic DNA was extracted from the ethanol preserved samples of the isolated parasites using QIAmp DNA Mini Kit (Qiagen, Germany) according to the manufacturer's instructions, and further stored at−20°C until used. PCR reactions were carried out using the following primers 25(28Sa F: 5′-TGCTTGTCTCAAAGATTAAGCC-3′, 28Sb R: 5′-TGCTTGTCTCAAAGATTAAGCC-3′). To target the 18S rRNA gene in a final reaction, a total volume of 50μl was prepared containing 1μl of DNA templates (100 ng), 0.5μl (50pmol) of each primer, 2μl of dNTPs (Advanced Bioenzymes, UK), 1μl DNA polymerase (2 U) (Advanced Bioenzymes, UK), 5μl of 10×buffer (500mM KCl, 100mM Tris HCl pH 9.0, 1.5mM MgCl2), and 40μl of dH2O. The temperature profile was as follows: Initial denaturation at 95°C for 5min, followed by a denaturation step of 35 cycles at 94°C for 45sec, annealing at 49°C for 45sec, extension at 72°C for 1min, and a final extension at 72°C for 1min. The amplification reactions were carried out in a PCR Thermocycler (PTC 100, MJR Research, USA). Two microliters of bromophenol blue were added to the aliquots of PCR products and the corresponding amplicons were electrophoresed on 2.5% agarose gel, stained with ethidium bromide, visualized using a UV transilluminator.
Sequencing and phylogenyThe PCR product was purified using a kit (Roche Diagnostics, Germany), and sequencing templates were prepared using a plasmid preparation kit (Machery–Nagel) and aBIO Dye Terminator v 3.1 Ready Sequencing Kit (Applied Biosystems) and 310 Automated DNA Sequencer (Applied Biosystems, USA) using the same primers. To identify related sequences, a BLAST search was carried out on the NCBI database. A BLAST rectangular tree was created from genetic distances calculated using the Jukes-Cantor model for nucleotide comparisons35 between the query sequence and arthropod sequences. The tree was built from the distance metrics using Fast Minimum Evolution (FastME) according to Desper and Gascuel16. Moreover, data of DNA sequences were aligned using CLUSTAL–X multiple sequence alignment58 and compared with previously deposited sequences of pentastomids recovered from GenBank to analyze intra–specific differences. The alignment was corrected manually using the alignment editor on BIOEDIT software 4.8.9. A phylogenetic tree was constructed using the MEGA program version 756.
ResultsAmong the 17 specimens of E. schneideri examined, twelve animals (prevalence 70.59%), eight males and four females, had their lungs infected by adult pentastomids. The infection increased during summer (4 males, 4 females) with a percentage of 89% and decreased during winter (4 males, 0 males) with a percentage of 57%. Adults were identified by the presence of tapered hooks devoid of chitinous accessories. The examined male hosts were generally more heavily infected (mean intensity of 4.3) than females (2.2).
MorphologyRaillietiella aegypti Ali et al.3Fig. 1 (A-G), Fig. 2 (A-H), Fig. 3 (A-H)
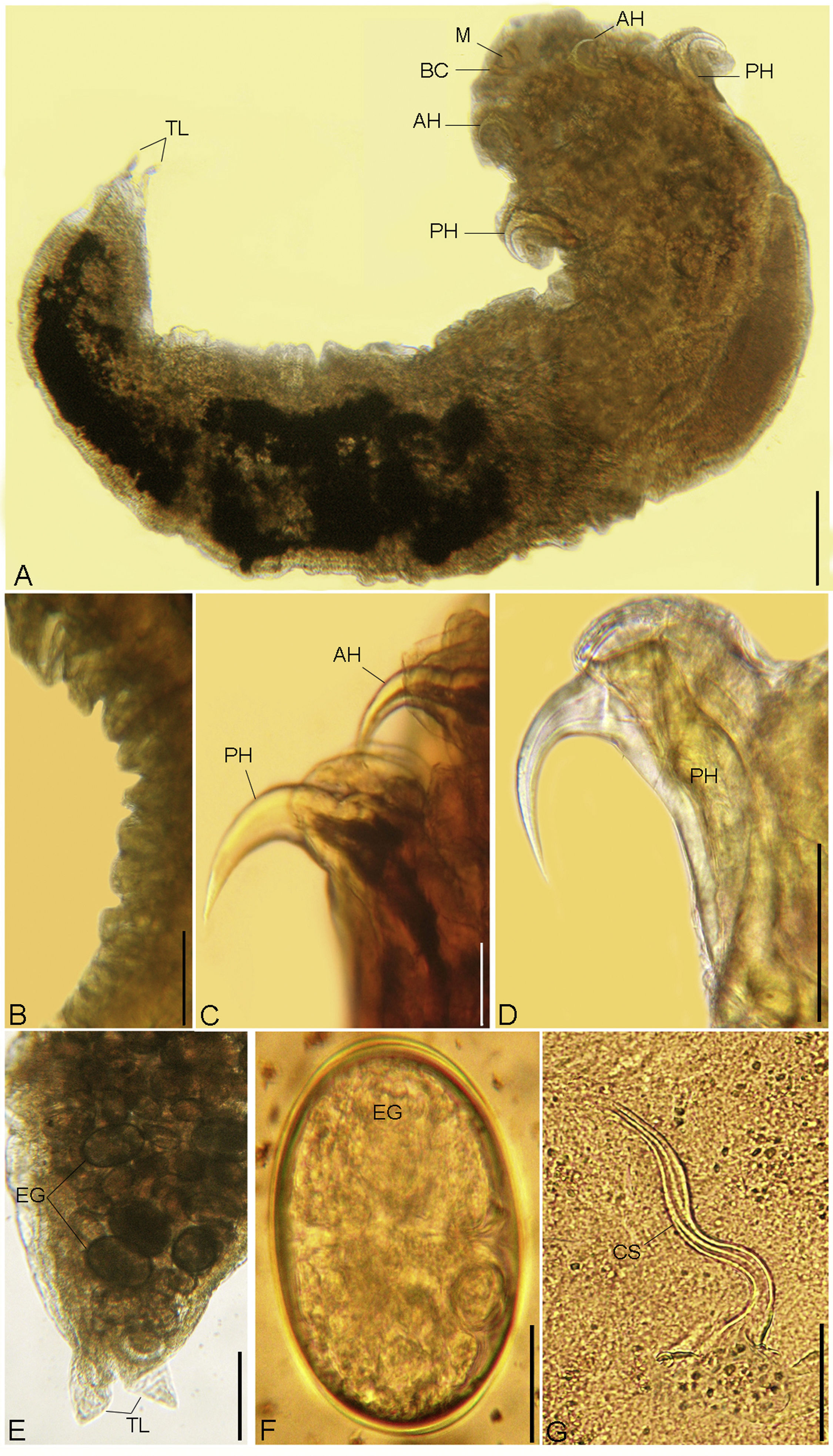
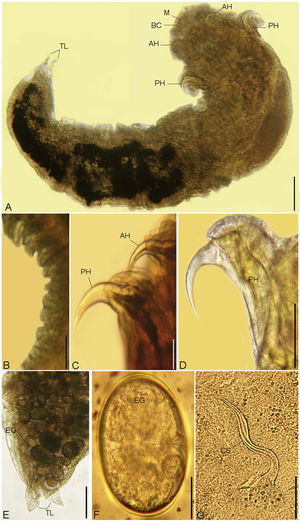
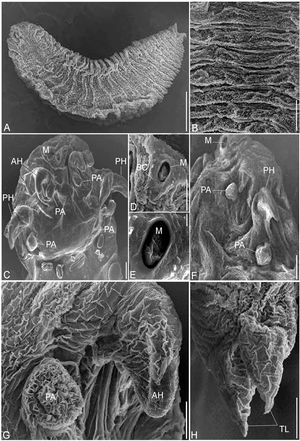
(A - G). Photomicrographs of R. aegypti showing: A Whole mount preparation of adult female with a broad cephalothorax that consists of a terminal mouth (M) at the apex supported by buccal cadre (BC), a pair of anterior hooks (AH), and a pair of sharp tipped posterior hooks (PH), abdomen terminated in divergent terminal lobes (TL), bar 50μm. B Abdominal annuli, bar 100μm. C, D Anterior (AH) and posterior (PH) hooks, bars 100μm. E Posterior part of a female abdomen with terminal lobes (TL), note the presence of eggs (EG), bar 50μm. F Eggs (EG), bar 10μm. G Copulatory spicule of male (CS), bar 10μm.
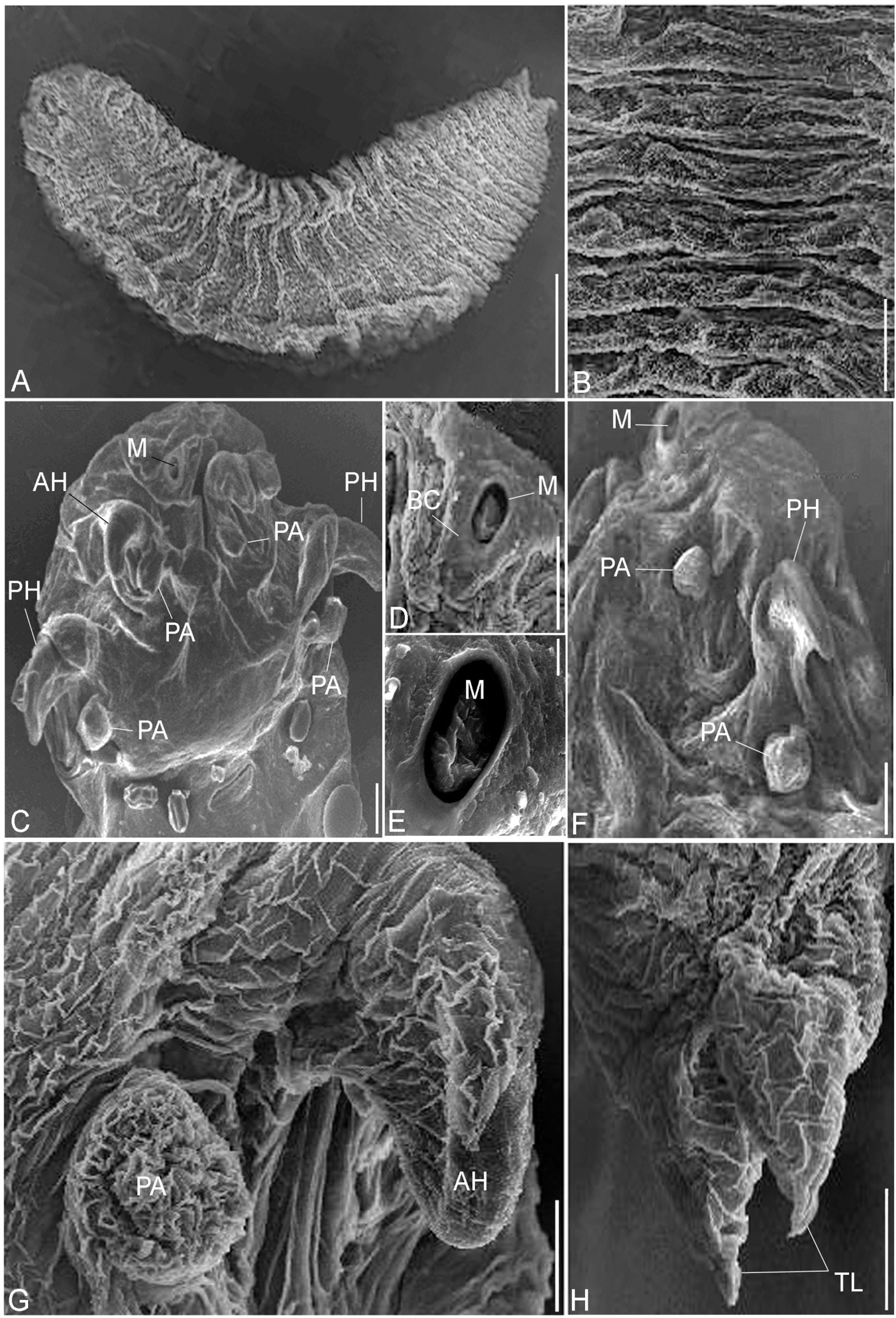
(A - H). Scanning electron micrographs of R. aegypti showing: A The adult parasite with an anterior cephalothorax and abdomen terminated in divergent terminal lobes, bar 500μm. B Abdominal annuli, bar 100μm. C Cephalothorax consists of a terminal mouth (M), two pairs of hooks, anterior (AH) and sharp posterior (PH), each with prominent cephalic papillae beside (PA), bar 100μm. D, E The terminal mouth surrounded by a buccal cadre (BC), bars 100μm; 10μm. F Lateral view, posterior hook (PS) and papillae (PA), bar 100μm. G Anterior hook (AH) and its papillae (PA), bar 50μm. H Abdomen terminated in divergent lobes surrounding anal opening, bar 100μm.
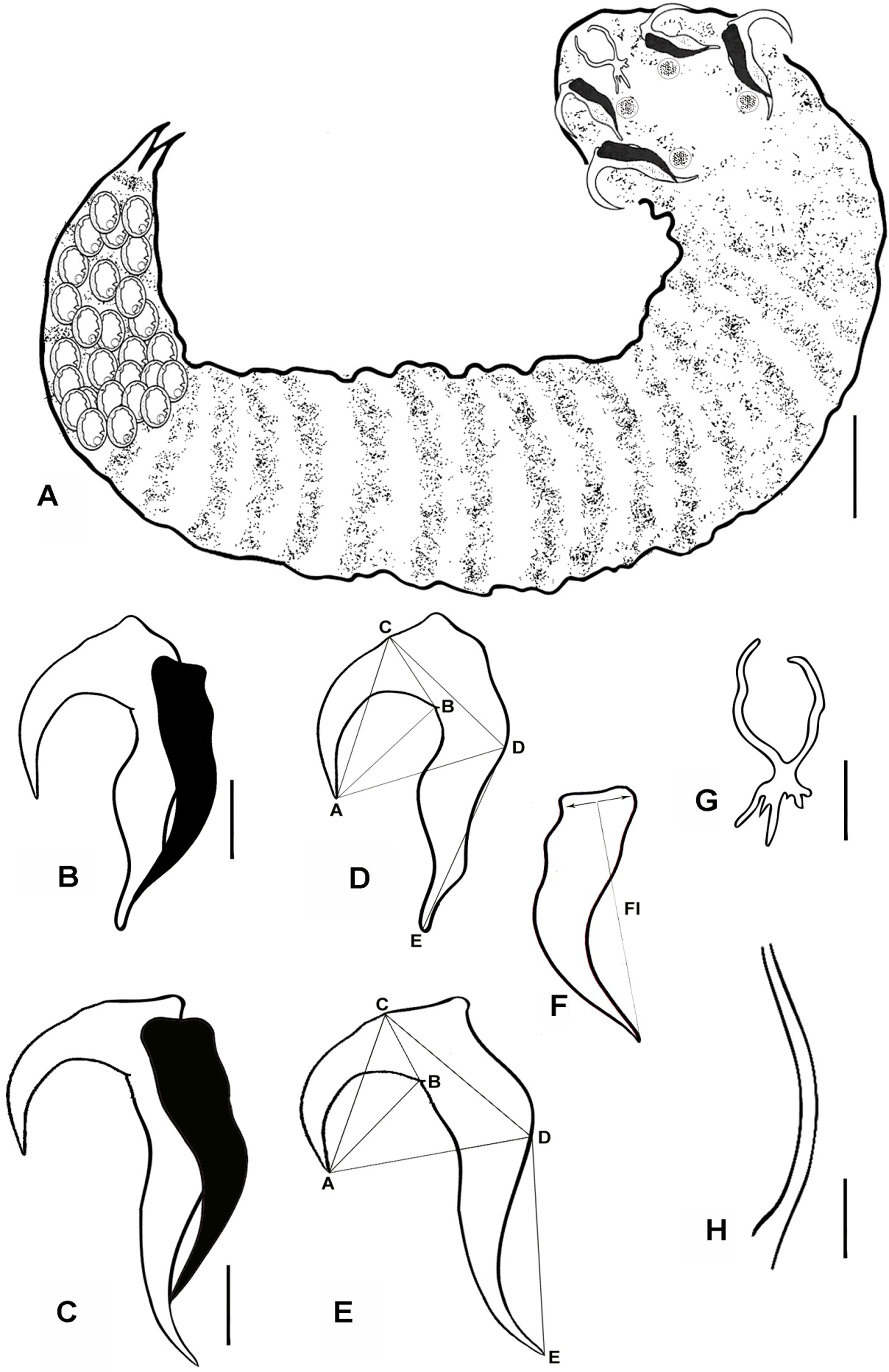
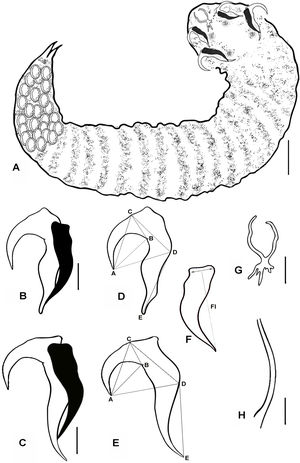
(A - H). Line diagrams showing: A Paratype female of R. aegypti with annuli, there are lobes and hooks in the area of cephalothorax, bar 50μm. B. Anterior hook with supporting fulcrum; C. Posterior hook with supporting fulcrum, bar 10μm. D, E, F. Hook measurement conventions. Anterior (D) and posterior (E) hooks and fulcrum (F) of an adult female, AC, Blade length; AD, Hook length; BC, Base length; CD, Plateau length; AB, Hook gape; DE Hook rest length; Fl, fulcrum length, measured from mid–point between anterior most lateral projections and posterior end, bar 10μm. G. Buccal cadre, bar 10μm. H. Copulatory spicule, bar 10μm.
Measurements and description based on the holotype female and male and paratype females.
Female (based on 5 specimens): body pyriform, annulated 25±2 (24–28), widened anteriorly between 3–14 annuli and tapered posteriorly to a bilobed terminal segment. Body was 16.22±4 (15.25–18.31) mm long and 4.16±0.21 (3.6–5.9) mm wide. Cephalothorax trapezoidal, slightly wider and indistinct from abdomen, rostrum not prominent, with a sclerotized U– shaped buccal cadre and a thick–framed terminal mouth. Two pairs of hooks, a pair of sharp anterior hooks with AB 135±5 (110–146) μm and BC 158±5 (150–169) μm and a pair of sharp posterior hooks with AB 221±5 (200–236) μm and BC 286±6 (280–289) μm. Four dorsal and prominent apical papillae present, one adjacent to each hook. Abdomen with distinct annuli, parapodial lobes present, anus ventral; caudal papillae indistinct; uterus straight, tube–like, and opened anteriorly.
Male (based on 5 specimens): body morphology was similar to that of the female; with a pyriform body, annulated 19±2 (16–21), widest anteriorly, 12.7±3 (10.5–13.3) mm long and 3.45±0.13 (3.2–5.8) mm wide, anterior hook with AB 73±3 (72–75) μm and BC 102±5 (100–103) μm; posterior hook larger with AB 190.6±5 (190–191) μm and BC 221±5 (280–289) μm. Dorsal papillae present; cephalothorax trapezoidal, widest at annuli 3–12, indistinct from abdomen. Two copulatory spicules curved with absolute uniform width; anus ventral; testis single, rounded, opened anteriorly.
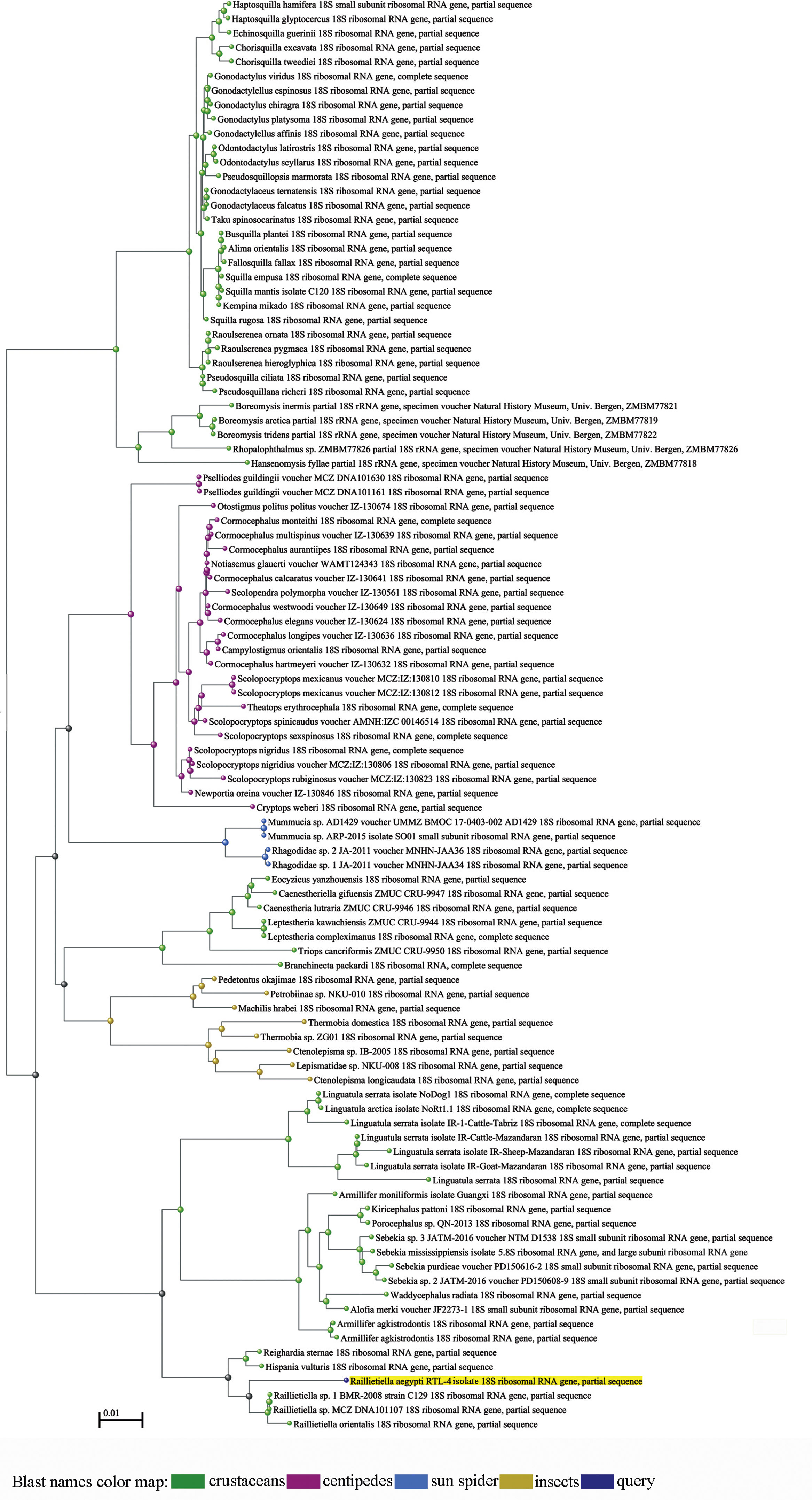
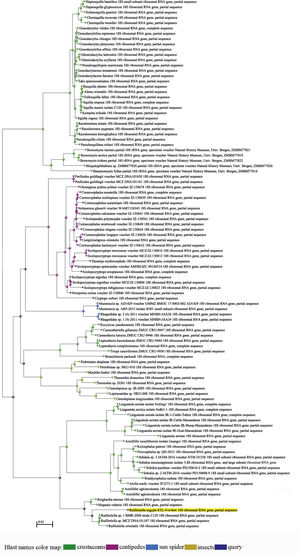
Molecular studySequence alignment by the maximum likelihood analysis for the data obtained from 18S rDNA analysis of the parasite isolated from the host examined yielded 1405bp, which exhibit identities ranging between 88–95% with pentastomid genera recovered from GenBank. BLAST computed a pairwise alignment between a query and the database sequences searched; a tree was constructed and constituted different clades from arthropod species (Figs. 4 and 5). It was observed that a separate monophyletic clade of Pentastomida was constructed including the present sequences and pentastomid species. Moreover, the results obtained strongly support that pentastomes are included within crustaceans and most closely related to branchiurans. The constructed phylogenetic tree by the maximum likelihood method based on the Tamura 3-parameter model among different species of the class Pentastomida arranged the aligned sequences and the present one into two main clades; the first clade (clade I) included species of the order Porocephalida34 while the second clade (clade II) included species of the order Cephalobaenida34 which was subdivided into two subclades: the first subclade (subclade A) arranged the species of family Reighardiida7: Hispania vulturis and Reighardiasternae19 exhibited 94.90% and 94.76 identities respectively with the query sequences. The second subclade (subclade B) arranged the species of family Raillietiellida7 to which the present sequences were included, the subclade B species and their identities being: R. orientalis31, 95.33% and Raillietiella sp. 25, 95.89% showed maximum identities with the query sequences. Estimates of Evolutionary Divergence between Sequences were shown in Table 1. The recovered sequences from the present study were deposited in GenBank under Accession number MK970649.1.
A rectangular tree drawn through the BLAST analysis. The terminal nodes are labeled by blast name to highlight taxonomic trends. The R. aegypti query sequence clusters with the arthropod sequences generated from the database sequences obtained from BLAST. The different blast names in the map distinguished by colors for crustaceans, centipedes, sun spider, insects and the query sequences.
Molecular phylogenetic analysis by the Maximum Likelihood method: The evolutionary history was inferred by using the Maximum Likelihood method based on the Tamura 3-parameter model. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree (s) for the heuristic search was/were obtained automatically by applying the Neighbor-Joining and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 23 nucleotide sequences. There were a total of 1396 positions in the final dataset. Evolutionary analyses were conducted in MEGA7.
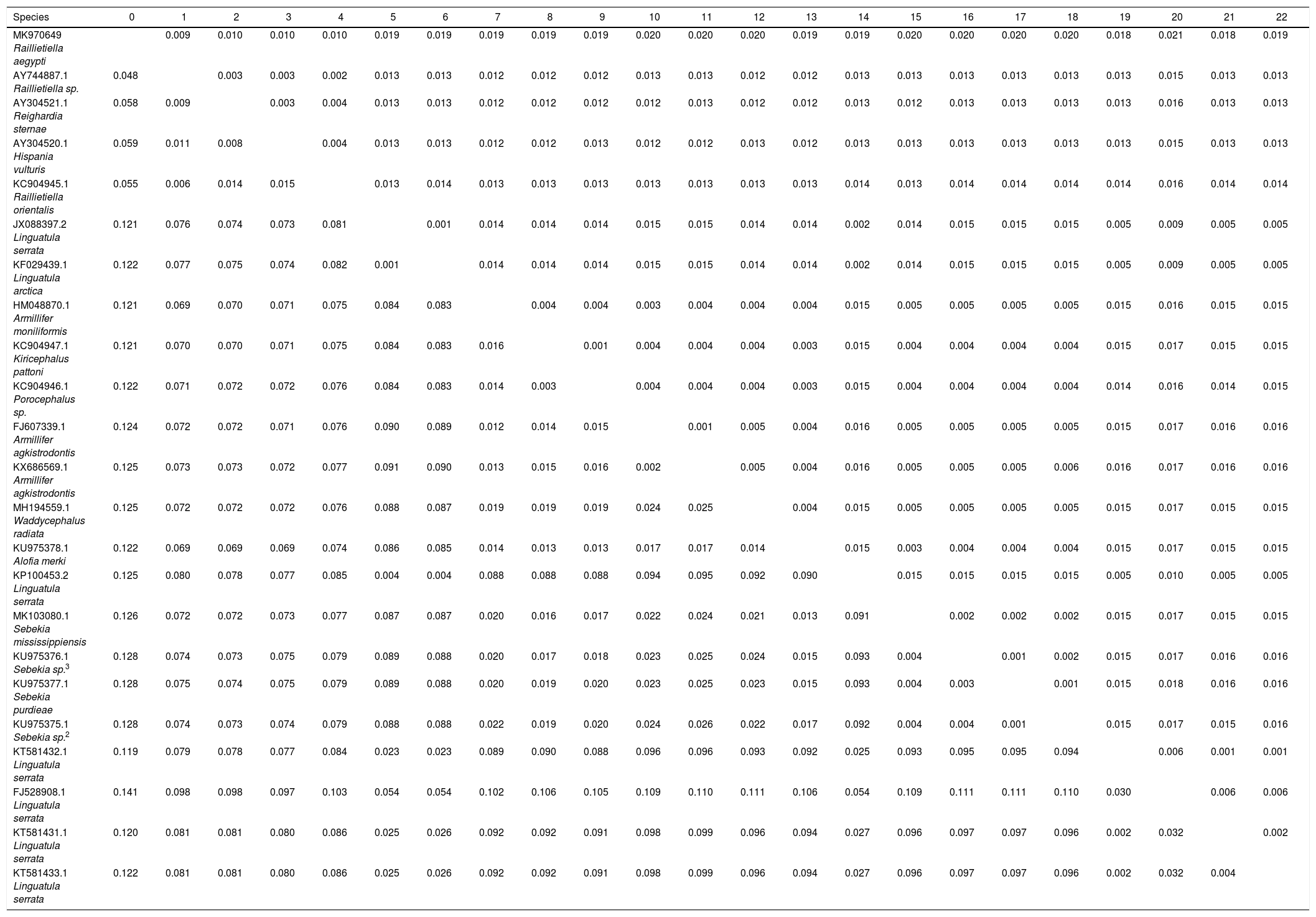
Estimates of Evolutionary Divergence between Sequences. The number of base substitutions per site from between sequences is shown. Standard error estimate (s) are shown above the diagonal. Analyses were conducted using the Maximum Composite Likelihood model. The analysis involved 23 nucleotide sequences. Codon positions included were 1st+2nd+3rd. All positions containing gaps and missing data were eliminated. There were a total of 1396 positions in the final dataset. Evolutionary analyses were conducted in MEGA7.
| Species | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MK970649 Raillietiella aegypti | 0.009 | 0.010 | 0.010 | 0.010 | 0.019 | 0.019 | 0.019 | 0.019 | 0.019 | 0.020 | 0.020 | 0.020 | 0.019 | 0.019 | 0.020 | 0.020 | 0.020 | 0.020 | 0.018 | 0.021 | 0.018 | 0.019 | |
| AY744887.1 Raillietiella sp. | 0.048 | 0.003 | 0.003 | 0.002 | 0.013 | 0.013 | 0.012 | 0.012 | 0.012 | 0.013 | 0.013 | 0.012 | 0.012 | 0.013 | 0.013 | 0.013 | 0.013 | 0.013 | 0.013 | 0.015 | 0.013 | 0.013 | |
| AY304521.1 Reighardia sternae | 0.058 | 0.009 | 0.003 | 0.004 | 0.013 | 0.013 | 0.012 | 0.012 | 0.012 | 0.012 | 0.013 | 0.012 | 0.012 | 0.013 | 0.012 | 0.013 | 0.013 | 0.013 | 0.013 | 0.016 | 0.013 | 0.013 | |
| AY304520.1 Hispania vulturis | 0.059 | 0.011 | 0.008 | 0.004 | 0.013 | 0.013 | 0.012 | 0.012 | 0.013 | 0.012 | 0.012 | 0.013 | 0.012 | 0.013 | 0.013 | 0.013 | 0.013 | 0.013 | 0.013 | 0.015 | 0.013 | 0.013 | |
| KC904945.1 Raillietiella orientalis | 0.055 | 0.006 | 0.014 | 0.015 | 0.013 | 0.014 | 0.013 | 0.013 | 0.013 | 0.013 | 0.013 | 0.013 | 0.013 | 0.014 | 0.013 | 0.014 | 0.014 | 0.014 | 0.014 | 0.016 | 0.014 | 0.014 | |
| JX088397.2 Linguatula serrata | 0.121 | 0.076 | 0.074 | 0.073 | 0.081 | 0.001 | 0.014 | 0.014 | 0.014 | 0.015 | 0.015 | 0.014 | 0.014 | 0.002 | 0.014 | 0.015 | 0.015 | 0.015 | 0.005 | 0.009 | 0.005 | 0.005 | |
| KF029439.1 Linguatula arctica | 0.122 | 0.077 | 0.075 | 0.074 | 0.082 | 0.001 | 0.014 | 0.014 | 0.014 | 0.015 | 0.015 | 0.014 | 0.014 | 0.002 | 0.014 | 0.015 | 0.015 | 0.015 | 0.005 | 0.009 | 0.005 | 0.005 | |
| HM048870.1 Armillifer moniliformis | 0.121 | 0.069 | 0.070 | 0.071 | 0.075 | 0.084 | 0.083 | 0.004 | 0.004 | 0.003 | 0.004 | 0.004 | 0.004 | 0.015 | 0.005 | 0.005 | 0.005 | 0.005 | 0.015 | 0.016 | 0.015 | 0.015 | |
| KC904947.1 Kiricephalus pattoni | 0.121 | 0.070 | 0.070 | 0.071 | 0.075 | 0.084 | 0.083 | 0.016 | 0.001 | 0.004 | 0.004 | 0.004 | 0.003 | 0.015 | 0.004 | 0.004 | 0.004 | 0.004 | 0.015 | 0.017 | 0.015 | 0.015 | |
| KC904946.1 Porocephalus sp. | 0.122 | 0.071 | 0.072 | 0.072 | 0.076 | 0.084 | 0.083 | 0.014 | 0.003 | 0.004 | 0.004 | 0.004 | 0.003 | 0.015 | 0.004 | 0.004 | 0.004 | 0.004 | 0.014 | 0.016 | 0.014 | 0.015 | |
| FJ607339.1 Armillifer agkistrodontis | 0.124 | 0.072 | 0.072 | 0.071 | 0.076 | 0.090 | 0.089 | 0.012 | 0.014 | 0.015 | 0.001 | 0.005 | 0.004 | 0.016 | 0.005 | 0.005 | 0.005 | 0.005 | 0.015 | 0.017 | 0.016 | 0.016 | |
| KX686569.1 Armillifer agkistrodontis | 0.125 | 0.073 | 0.073 | 0.072 | 0.077 | 0.091 | 0.090 | 0.013 | 0.015 | 0.016 | 0.002 | 0.005 | 0.004 | 0.016 | 0.005 | 0.005 | 0.005 | 0.006 | 0.016 | 0.017 | 0.016 | 0.016 | |
| MH194559.1 Waddycephalus radiata | 0.125 | 0.072 | 0.072 | 0.072 | 0.076 | 0.088 | 0.087 | 0.019 | 0.019 | 0.019 | 0.024 | 0.025 | 0.004 | 0.015 | 0.005 | 0.005 | 0.005 | 0.005 | 0.015 | 0.017 | 0.015 | 0.015 | |
| KU975378.1 Alofia merki | 0.122 | 0.069 | 0.069 | 0.069 | 0.074 | 0.086 | 0.085 | 0.014 | 0.013 | 0.013 | 0.017 | 0.017 | 0.014 | 0.015 | 0.003 | 0.004 | 0.004 | 0.004 | 0.015 | 0.017 | 0.015 | 0.015 | |
| KP100453.2 Linguatula serrata | 0.125 | 0.080 | 0.078 | 0.077 | 0.085 | 0.004 | 0.004 | 0.088 | 0.088 | 0.088 | 0.094 | 0.095 | 0.092 | 0.090 | 0.015 | 0.015 | 0.015 | 0.015 | 0.005 | 0.010 | 0.005 | 0.005 | |
| MK103080.1 Sebekia mississippiensis | 0.126 | 0.072 | 0.072 | 0.073 | 0.077 | 0.087 | 0.087 | 0.020 | 0.016 | 0.017 | 0.022 | 0.024 | 0.021 | 0.013 | 0.091 | 0.002 | 0.002 | 0.002 | 0.015 | 0.017 | 0.015 | 0.015 | |
| KU975376.1 Sebekia sp.3 | 0.128 | 0.074 | 0.073 | 0.075 | 0.079 | 0.089 | 0.088 | 0.020 | 0.017 | 0.018 | 0.023 | 0.025 | 0.024 | 0.015 | 0.093 | 0.004 | 0.001 | 0.002 | 0.015 | 0.017 | 0.016 | 0.016 | |
| KU975377.1 Sebekia purdieae | 0.128 | 0.075 | 0.074 | 0.075 | 0.079 | 0.089 | 0.088 | 0.020 | 0.019 | 0.020 | 0.023 | 0.025 | 0.023 | 0.015 | 0.093 | 0.004 | 0.003 | 0.001 | 0.015 | 0.018 | 0.016 | 0.016 | |
| KU975375.1 Sebekia sp.2 | 0.128 | 0.074 | 0.073 | 0.074 | 0.079 | 0.088 | 0.088 | 0.022 | 0.019 | 0.020 | 0.024 | 0.026 | 0.022 | 0.017 | 0.092 | 0.004 | 0.004 | 0.001 | 0.015 | 0.017 | 0.015 | 0.016 | |
| KT581432.1 Linguatula serrata | 0.119 | 0.079 | 0.078 | 0.077 | 0.084 | 0.023 | 0.023 | 0.089 | 0.090 | 0.088 | 0.096 | 0.096 | 0.093 | 0.092 | 0.025 | 0.093 | 0.095 | 0.095 | 0.094 | 0.006 | 0.001 | 0.001 | |
| FJ528908.1 Linguatula serrata | 0.141 | 0.098 | 0.098 | 0.097 | 0.103 | 0.054 | 0.054 | 0.102 | 0.106 | 0.105 | 0.109 | 0.110 | 0.111 | 0.106 | 0.054 | 0.109 | 0.111 | 0.111 | 0.110 | 0.030 | 0.006 | 0.006 | |
| KT581431.1 Linguatula serrata | 0.120 | 0.081 | 0.081 | 0.080 | 0.086 | 0.025 | 0.026 | 0.092 | 0.092 | 0.091 | 0.098 | 0.099 | 0.096 | 0.094 | 0.027 | 0.096 | 0.097 | 0.097 | 0.096 | 0.002 | 0.032 | 0.002 | |
| KT581433.1 Linguatula serrata | 0.122 | 0.081 | 0.081 | 0.080 | 0.086 | 0.025 | 0.026 | 0.092 | 0.092 | 0.091 | 0.098 | 0.099 | 0.096 | 0.094 | 0.027 | 0.096 | 0.097 | 0.097 | 0.096 | 0.002 | 0.032 | 0.004 |
Studying parasite diversity worldwide is important for at least two major reasons. First, parasites are now recognized as playing important roles in ecosystem fractions by influencing the populations and communities of their hosts39. Second, many parasite species are agriculturally and medically important15,26,43. Therefore, the identification of parasitic worms based on microscopic observations and PCR amplification/sequencing of 18S rDNA from isolated parasites is highly recommended for successful investigations and taxonomy44. There are many parasitic crustaceans, such as pentastomes, whose adult morphology is devoid of crustacean features.
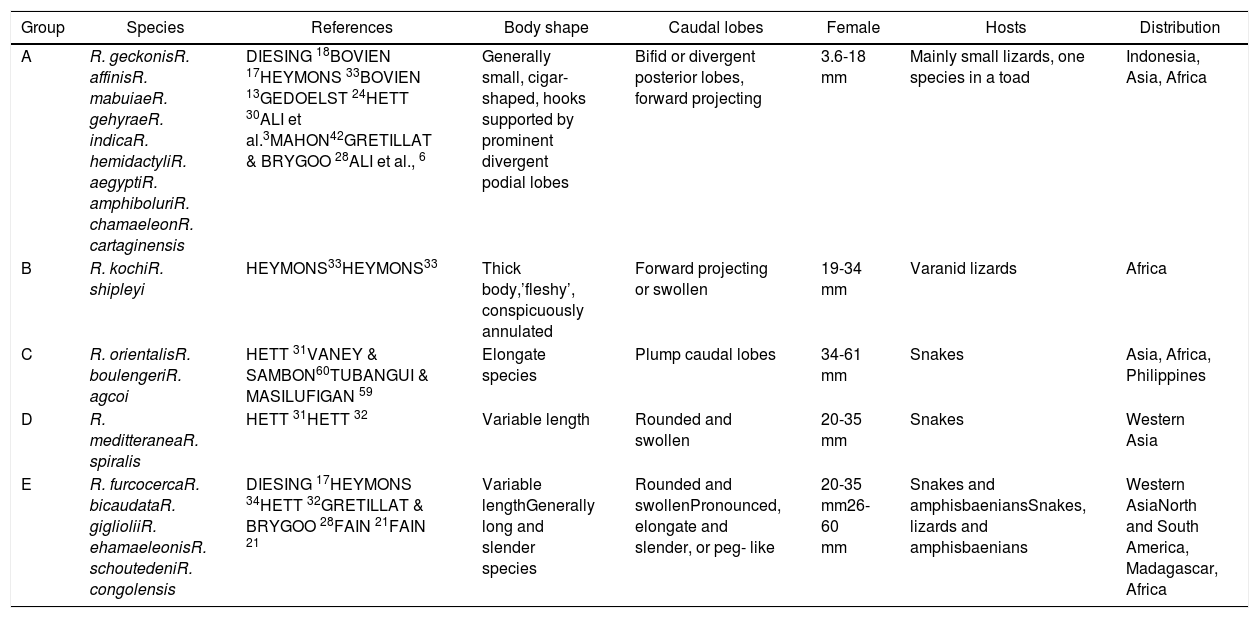
Family Raillietiellidae represented by the single pentastomid genus Raillietiella is characterized by a quite wide diversity of hosts7,8. The taxonomy and systematics of the pentastomid genus Raillietiella were reviewed by Ali et al.6 in the description of R. cartagenensis and in the redescription of R. amphiboluri42, R. kochi33, R. shipleyi33, and R. indica24. Ali et al.6 reorganized the known and valid species of Raillietiella into five groups (Table 2): small lizards, varanid lizards, amphisbaenians, snakes, and amphibians, which are mainly based on host characteristics (host type, ecology, and zoogeography)50. Two of these groups (groups I and II; after Ali et al.6) include species infecting small insectivorous lizards and are easily differentiated by the initial ideas of Self55 into species with sharp-tipped posterior hooks (group I) and blunt-tip posterior hooks (group II). Species differentiation in these two groups is mainly based on a combination of characteristics, including body size, annulus number, posterior-hook dimensions, and size and shape of the male copulatory spicule50. The sharp-tipped posterior-hook of Raillietiella spp. includes six well-characterized species: R. amphiboluri42 infecting the Australian bearded lizard Amphibolurus barbatus; R. chamaeleonis28 in Chamaeleo oustaleti and Chamaeleo verrucosus from Madagascar; R. mottae9 from Tropidurus hispidus from Northeastern Brazil; R. morenoi1 from Gallotia atlantica in the Canary Islands, R. aegypti3 in different small lizards from Egypt; and R. cartagenensis6 in Hemidactylus sp. and Gonatodes sp. from Colombia. The reported presence of R. affinis13 in Lepidactylus lugutris from the British Salomon Island still awaits confirmation and also that in Lioheterodon modestus from Madagascar due to the inclusion of R. chamaeleonis as a host27. The present Raillietiella sp. could be distinguished from Raillietilla sharp-hooked species of small lizards through different combinations of characteristics (Table 3), such as measurements of body length, annulus number, anterior-hook and posterior-hook size, and dimensions of the male copulatory spicule. Furthermore, it can be differentiated from similar species in group I. The taxonomic and systematic morphological characteristics stated by Ali et al.6 and others referring to male hooks50 are reliable in distinguishing and separating the present Raillietiella sp. from the rest. Based on light and scanning electron microscopy, the present parasite should be classified morphologically as R. aegypti3 according to the following criteria: the same host with a new type locality; it is a group I type of Raillietiella spp. and the dimensions of anterior and posterior hook. This is the first report of a pentastomid in a reptile species from Saudi Arabia. The congruence between molecular and morphological data in the taxonomic justification of pentastomids was demonstrated by Mohanta and Itagaki44.
The five groups of Raillietiellids modified from Ali et al.6
| Group | Species | References | Body shape | Caudal lobes | Female | Hosts | Distribution |
|---|---|---|---|---|---|---|---|
| A | R. geckonisR. affinisR. mabuiaeR. gehyraeR. indicaR. hemidactyliR. aegyptiR. amphiboluriR. chamaeleonR. cartaginensis | DIESING 18BOVIEN 17HEYMONS 33BOVIEN 13GEDOELST 24HETT 30ALI et al.3MAHON42GRETILLAT & BRYGOO 28ALI et al., 6 | Generally small, cigar- shaped, hooks supported by prominent divergent podial lobes | Bifid or divergent posterior lobes, forward projecting | 3.6-18 mm | Mainly small lizards, one species in a toad | Indonesia, Asia, Africa |
| B | R. kochiR. shipleyi | HEYMONS33HEYMONS33 | Thick body,’fleshy’, conspicuously annulated | Forward projecting or swollen | 19-34 mm | Varanid lizards | Africa |
| C | R. orientalisR. boulengeriR. agcoi | HETT 31VANEY & SAMBON60TUBANGUI & MASILUFIGAN 59 | Elongate species | Plump caudal lobes | 34-61 mm | Snakes | Asia, Africa, Philippines |
| D | R. meditteraneaR. spiralis | HETT 31HETT 32 | Variable length | Rounded and swollen | 20-35 mm | Snakes | Western Asia |
| E | R. furcocercaR. bicaudataR. giglioliiR. ehamaeleonisR. schoutedeniR. congolensis | DIESING 17HEYMONS 34HETT 32GRETILLAT & BRYGOO 28FAIN 21FAIN 21 | Variable lengthGenerally long and slender species | Rounded and swollenPronounced, elongate and slender, or peg- like | 20-35 mm26-60 mm | Snakes and amphisbaeniansSnakes, lizards and amphisbaenians | Western AsiaNorth and South America, Madagascar, Africa |
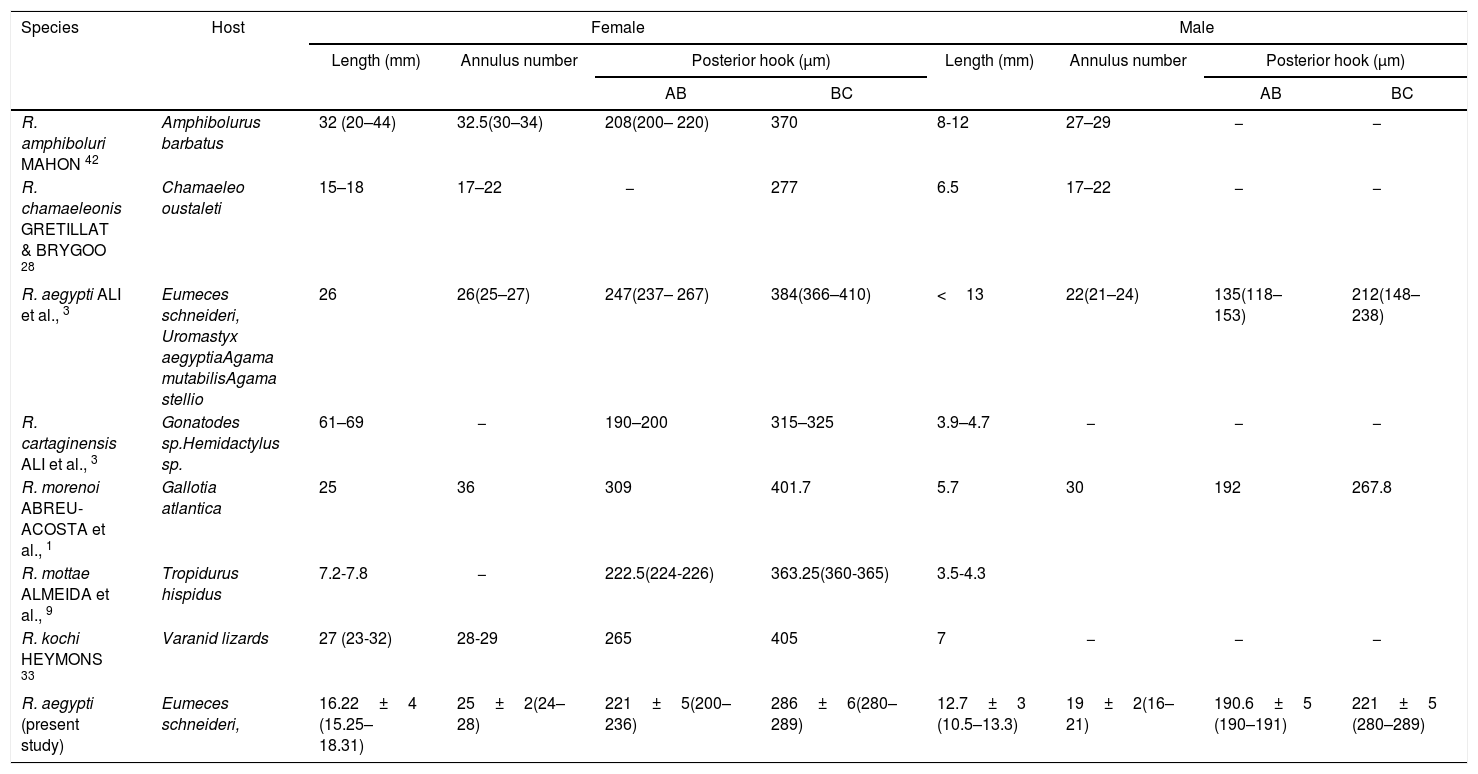
A summary of the principal characteristics of sharp-hooked Raillietiella species from insectivorous lizards.
| Species | Host | Female | Male | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Length (mm) | Annulus number | Posterior hook (μm) | Length (mm) | Annulus number | Posterior hook (μm) | ||||
| AB | BC | AB | BC | ||||||
| R. amphiboluri MAHON 42 | Amphibolurus barbatus | 32 (20–44) | 32.5(30–34) | 208(200– 220) | 370 | 8-12 | 27–29 | − | − |
| R. chamaeleonis GRETILLAT & BRYGOO 28 | Chamaeleo oustaleti | 15–18 | 17–22 | − | 277 | 6.5 | 17–22 | − | − |
| R. aegypti ALI et al., 3 | Eumeces schneideri, Uromastyx aegyptiaAgama mutabilisAgama stellio | 26 | 26(25–27) | 247(237– 267) | 384(366–410) | <13 | 22(21–24) | 135(118–153) | 212(148–238) |
| R. cartaginensis ALI et al., 3 | Gonatodes sp.Hemidactylus sp. | 61–69 | − | 190–200 | 315–325 | 3.9–4.7 | − | − | − |
| R. morenoi ABREU-ACOSTA et al., 1 | Gallotia atlantica | 25 | 36 | 309 | 401.7 | 5.7 | 30 | 192 | 267.8 |
| R. mottae ALMEIDA et al., 9 | Tropidurus hispidus | 7.2-7.8 | − | 222.5(224-226) | 363.25(360-365) | 3.5-4.3 | |||
| R. kochi HEYMONS 33 | Varanid lizards | 27 (23-32) | 28-29 | 265 | 405 | 7 | − | − | − |
| R. aegypti (present study) | Eumeces schneideri, | 16.22±4 (15.25–18.31) | 25±2(24–28) | 221±5(200–236) | 286±6(280–289) | 12.7±3 (10.5–13.3) | 19±2(16–21) | 190.6±5 (190–191) | 221±5 (280–289) |
Our analysis corroborates the monophyly of Pentastomida and the results thus provide strong support for Wingstrand's proposal that pentastomes are highly modified crustaceans most closely related to branchiurans. Data derived from the DNA analysis coincide with those from morphological studies in accordance with Wägele61 and Nielsen45; the maximum identity obtained for the present sequences was 95.89% with Raillietiella sp. recovered from different hosts and localities with some morphological differences that exclude the suggestion to classify the present species as being the same. Moreover, since the present parasite is morphologically the same as R. aegypti described previously with no molecular confirmation, the present study represents the first one to confirm the taxonomy of R. aegypti. The results of molecular analyses vary according to the selected species, sample size, and particular methods of analysis used, where the sequences of 18S rRNA may contain consistent phylogenetic information for cladogenetic events as old as the Median Cambrian9,47. This morphology–based taxonomy is consistent with present molecular phylogenies, which are supported by previous observations. Consequently, revision on the morphology–based taxonomy or inclusion of molecular data from an extensive range of taxa should be necessary to elucidate the phylogenetic relationships of the Pentastomida.
Animal Rights StatementAuthors declare that the experiments on animals were conducted in accordance with local Ethical Committee laws and regulations as regards care and use of laboratory animals.
Financial Disclosure StatementThe Deanship of Scientific Research at King Khalid University through Research group Project under grant number (R.G.P.1–112 –40).
Conflict of Interests StatementNo conflict of interests for the publication of this article was reported.
The authors extend their appreciation to the Deanship of Scientific Research at King Khalid University for funding this work through Research group Project under grant number (R.G.P.1–112 –40).