It has been recently found that the natural distribution, habitat, and genetic diversity of astaxanthin-producing yeasts (i.e. Phaffia rhodozyma, synonym Xanthophyllomyces dendrorhous) is much greater than previously thought. P. rhodozyma is biotechnologically exploited due to its ability to produce the carotenoid pigment astaxanthin and thus, it is used as a natural source of this pigment for aquaculture. P. rhodozyma was also capable of synthesizing the potent UVB sunscreen mycosporine-glutaminol-glucoside (MGG). Therefore, further environmental studies are needed to elucidate its ecological aspects and detect new potential strains for the production of astaxanthin and MGG. However, obtaining new isolates of P. rhodozyma and related species is not always easy due to its low abundance and the presence of other sympatric and pigmented yeasts. In this work we report a successful development of a species-specific primer which has the ability to quickly and accurately detecting isolates representing all known lineages of the genus Phaffia (including novel species of the genus) and excluding closely related taxa. For this purpose, a primer of 20 nucleotides (called PhR) was designed to be used in combination with universal primers ITS3 and NL4 in a multiplex amplification. The proposed method has the sensitivity and specificity required for the precise detection of new isolates, and therefore represents an important tool for the environmental search for novel astaxanthin-producing yeasts.
Recientemente, se ha encontrado que la distribución natural, el hábitat y la diversidad genética de levaduras productoras de astaxantina (p. ej., Phaffia rhodozyma, sinónimo Xanthophyllomyces dendrorhous) son mucho mayores de lo que se pensaba. P. rhodozyma se explota biotecnológicamente debido a su capacidad para producir el pigmento carotenoide astaxantina y, por lo tanto, se utiliza como una fuente natural de este pigmento para la acuicultura. También se encontró que esta levadura es capaz de sintetizar el potente protector solar UVB micosporina-glutaminol-glucósido (MGG). Por lo tanto, más estudios ambientales para dilucidar sus aspectos ecológicos y detectar nuevas cepas potenciales productoras de astaxantina y MGG son necesarios. Sin embargo, la obtención de nuevos aislamientos de P. rhodozyma y especies relacionadas no siempre es fácil debido a su baja abundancia y a la presencia de otras levaduras simpátricas y pigmentadas. En este trabajo se describe el desarrollo exitoso de un cebador especie-específico que tiene la capacidad de detectar rápidamente y con precisión cepas representativas de todos los linajes del género Phaffia previamente reportados (incluyendo nuevas especies del género) y excluir especies estrechamente relacionadas. Para ello, se diseñó un cebador de 20 nucleótidos (denominado PhR) para ser utilizado en combinación con los cebadores universales ITS3 y NL4 en una amplificación multiplex. El método propuesto tiene la sensibilidad y la especificidad requerida para la detección precisa de nuevos aislamientos y, por lo tanto, representa una importante herramienta para la búsqueda ambiental de nuevas levaduras productoras de astaxantina.
Phaffia rhodozyma (also known as Xanthophyllomyces dendrorhous) is a basidiomycetous yeast, forming orange-red colonies and known worldwide since, until now, it is the only yeast capable of producing the carotenoid pigment astaxanthin (3,3′-dihydroxy-β,β-carotene-4,4′-dione). This compound is of economic importance because it is the most expensive feed component for aquaculture and aviculture8. Moreover, P. rhodozyma is the only known carotenogenic yeast species that is able to vigorously ferment a number of sugars, including glucose, maltose, sucrose and raffinose9. Moreover, it has recently been discovered that this yeast has the ability to synthesize a UV-absorbing molecule called mycosporine-glutaminol-glucoside of applied interest14.
The natural distribution and habitat of P. rhodozyma are much broader than previously thought. Initially, genetically similar isolates of this yeast were found in the Northern Hemisphere in association with slime exudates of trees, being obtained from Japan, Canada, Russia, Italy, Germany and USA3,22,27,30. A novel and genetically distinct Phaffia population was later isolated in the Southern Hemisphere, from the sugary stromata of the Cyttaria hariotti fungus, a parasite of the Nothofagus trees in Argentina15. An even more genetically divergent strain was described in Chile, which might represent a novel species although a single strain is known29. More recently, David-Palma et al.2 investigated the association of Phaffia with Nothofagus-Cyttaria in Australasia, the other region of the world where Nothofagus are endemic, and discovered an even higher Phaffia diversity, including two endemic and markedly divergent linages which represent putative new species, both with the capability to produce astaxanthin and MGG (unpublished results). In this work it was suggested that Phaffia adaptation to different tree hosts/niches has driven population structure, thus, it can be anticipated that novel lineages of this yeast might be yet undiscovered. Proof of this fact are the recent isolates found by Yurkov et al.33, and Contreras et al.1 from soil and Antarctic-related environments, respectively. In this scenario and due to the biotechnological relevance of this genus, specific culture media and biochemical techniques that allow a more favorable isolation and simpler identification of P. rhodozyma were reported in previous works2,13,26. This is particularly important considering that the environmental isolation of P. rhodozyma is not always easy due to its low abundance and because it is easily misidentified with other sympatric and carotenoid-accumulating yeasts of the genera Rhodotorula, Cystofilobasidium and Dioszegia16. Not many yeast species are able to produce carotenoid pigments and these are distributed among a few lineages18. Those species accumulating mainly xanthophyll-like carotenoids that produce orange-type colonies (Cystofilobasidium and Dioszegia) have higher probabilities of misidentification with astaxanthin-producing yeasts like Phaffia. Other sympatric species such as many Rhodotorula spp. or few Cryptococcus spp. can produce salmon-pink or intense red colonies depending on the relative level of accumulation of less polar carotenoids like torulene, torularhodine and/or β-carotene18.
The objective of this work was to provide a rapid and accurate PCR-based detection method for astaxanthin-accumulating yeast isolates from environmental samples.
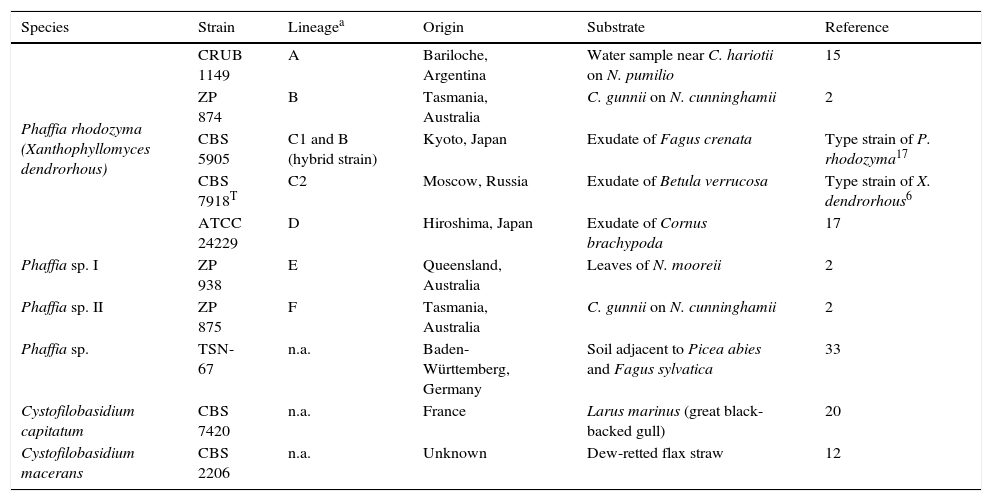
Materials and methodsYeast strainsSeven representative strains of the different Phaffia spp. clades described in David-Palma et al.2 were used in this study (Table 1). Strains ZP938 and ZP875 were kindly provided by Dr. J. Sampaio (UNL, Portugal). The Phaffia-related strain TSN-67 (astaxanthin and MGG non-producing, unpublished results) recently reported by Yurkov et al.33 was kindly provided by the author (A. Yurkov) and was tested together with two species of the genus Cystofilobasidium. The latter species were included as controls due to their phylogenetic proximity to P. rhodozyma (also belonging to the order Cystofilobasidiales), habitat co-occurrence and similar appearance of their colonies.
List of yeast strains tested with P. rhodozyma specific primers
| Species | Strain | Lineagea | Origin | Substrate | Reference |
|---|---|---|---|---|---|
| Phaffia rhodozyma (Xanthophyllomyces dendrorhous) | CRUB 1149 | A | Bariloche, Argentina | Water sample near C. hariotii on N. pumilio | 15 |
| ZP 874 | B | Tasmania, Australia | C. gunnii on N. cunninghamii | 2 | |
| CBS 5905 | C1 and B (hybrid strain) | Kyoto, Japan | Exudate of Fagus crenata | Type strain of P. rhodozyma17 | |
| CBS 7918T | C2 | Moscow, Russia | Exudate of Betula verrucosa | Type strain of X. dendrorhous6 | |
| ATCC 24229 | D | Hiroshima, Japan | Exudate of Cornus brachypoda | 17 | |
| Phaffia sp. I | ZP 938 | E | Queensland, Australia | Leaves of N. mooreii | 2 |
| Phaffia sp. II | ZP 875 | F | Tasmania, Australia | C. gunnii on N. cunninghamii | 2 |
| Phaffia sp. | TSN-67 | n.a. | Baden-Württemberg, Germany | Soil adjacent to Picea abies and Fagus sylvatica | 33 |
| Cystofilobasidium capitatum | CBS 7420 | n.a. | France | Larus marinus (great black-backed gull) | 20 |
| Cystofilobasidium macerans | CBS 2206 | n.a. | Unknown | Dew-retted flax straw | 12 |
C., Cyttaria; N., Nothofagus; n.a., Not applicable.
Phaffia spp. specific primer was designed based on concatenated sequences of internal transcribed spacers (ITS) and D1D2 partial sequences of 20 P. rhodozyma strains (available at GenBank by May, 2011) and 5 Cystofilobasidium spp. Using the NCBI primer BLAST tool and then checked using Primer3. Both widely used ITS3 and NL4 and new Phaffia spp. specific primers are shown with full detail in Table 2.
Phylogenetic analysisrRNA gene sequences corresponding to the internal transcribed spacers 1 and 2 (ITS 1 and ITS 2), 5.8S rRNA of all the strains depicted in Table 1 were obtained from NCBI. DNA sequences alignment and analyses were performed using MEGA 5.024. Alignments were carried out using CLUSTALW25 and were edited manually when required. Phylogenetic analysis using Neighbor Joining were performed in MEGA 5.0, using the Kimura 2-parameter model as substitution model. The reliability of the NJ trees was assessed by bootstrap analysis including 1000 replications.
Cell and molecular methodsCell growth was performed on YMA agar plates (3g/l yeast extract, 3g/l malt extract, 5g/l bactopeptone, 10g/l glucose and 15g/l agar), DNA was extracted using the protocol described previously in Libkind et al.11 and purified using chloroform:isoamyl alcohol reagent (24:1, v/v). DNA was quantified using Shimadzu UV-1800 spectrophotometer and Multiplex PCR assays were performed in a Labnet MultiGene Gradient thermocycler, under the following conditions: denaturation at 95°C for 5min followed by 35 cycles of 95°C 30s, 55°C 1min, 72°C 1min, and a final extension at 72°C for 7min. PCR amplification was performed in a total volume of 25μl, containing about 50ng of total DNA, 1× GoTaq PCR buffer (Promega), 200μM each dNTP (GE Healthcare), 1U Taq polymerase (GoTaq, Promega) and 0.3μM each primer. Gel electrophoresis was performed on 1% agarose in 0.5× TBE, 90V for 30min.
A 100bp DNA size ladder (Highway) plus a lambda DNA digested with HindIII (Promega) were used as a molecular marker.
ResultsA single primer of 20 nucleotides called PhR (Table 2) was designed to be used in combination with the universal primers ITS331 and NL410 to perform a multiplex amplification reaction that leads to the rapid detection of astaxanthin-producing yeasts of the genus Phaffia. The ability of this method to detect all proposed lineages of Phaffia sp. and at the same time retain the sufficient specificity to avoid false positives was evaluated. For this purpose, an up-to- date set of eight representative strains of all known Phaffia and Phaffia-related lineages were assayed, including the Phaffia sp. TSN-67 isolate reported by Yurkov et al.33 from soil and representative strains of the seven different lineages recently described by David-Palma et al2. Additionally, Cystofilobasidium capitatum and Cystofilobasidium macerans were included due to their phylogenetic proximity and, since they inhabit similar substrates and grow in morphologically similar colonies, they can be frequently confused with Phaffia4,5,22,27,28.
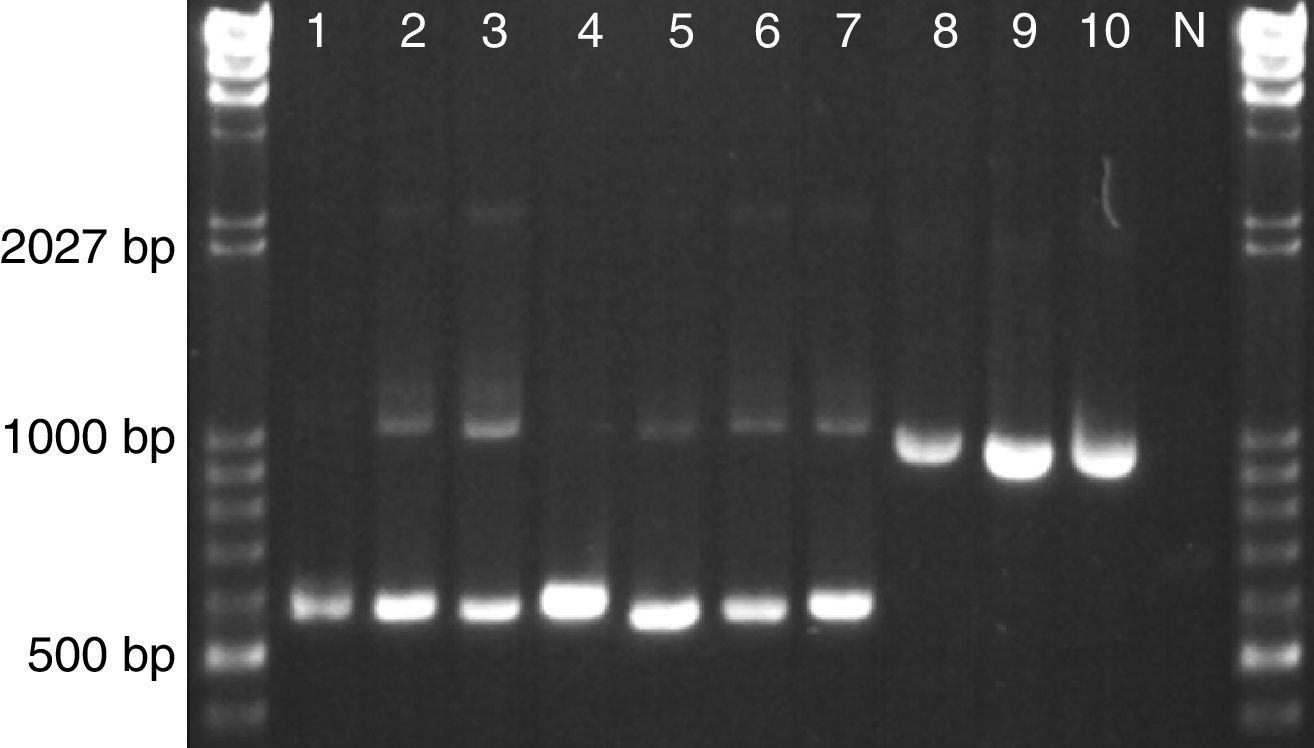
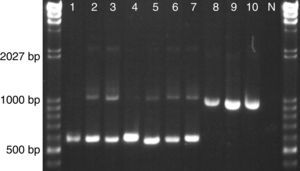
The reverse PhR primer was designed to anneal in the conserved D1D2 domains of the 26S rRNA gene and to generate a 643bp amplicon when it is used together with the universal forward primer ITS3. The universal reverse primer NL4 was included for amplification control since it will produce 1155bp in any eukaryotic microorganism. When tested in our set of astaxanthin-producing strains we observed the specific Phaffia band (∼640bp) for all P. rhodozyma lineages and even for strains of group E and F, which represent novel species of the genus Phaffia (Fig. 1). In most of these cases, the control band (∼1150bp) was also present though displaying a weak signal.
Yeast strains tested with multiplex PhR, ITS3 and NL4. 1–7 P. rhodozyma: (1) CBS 5905 (Lineage C1/B); (2) CBS 7918T (Lineage C2); (3) CRUB 1149 (Lineage A); (4) ATCC 24229 (Lineage D); (5) ZP 938 (Lineage E); (6) ZP 875 (Lineage F); (7) ZP 874 (Lineage B); (8) TSN-67; (9) C. capitatum CBS 7420; (10) C. macerans CBS 2206; N – negative control (no template).
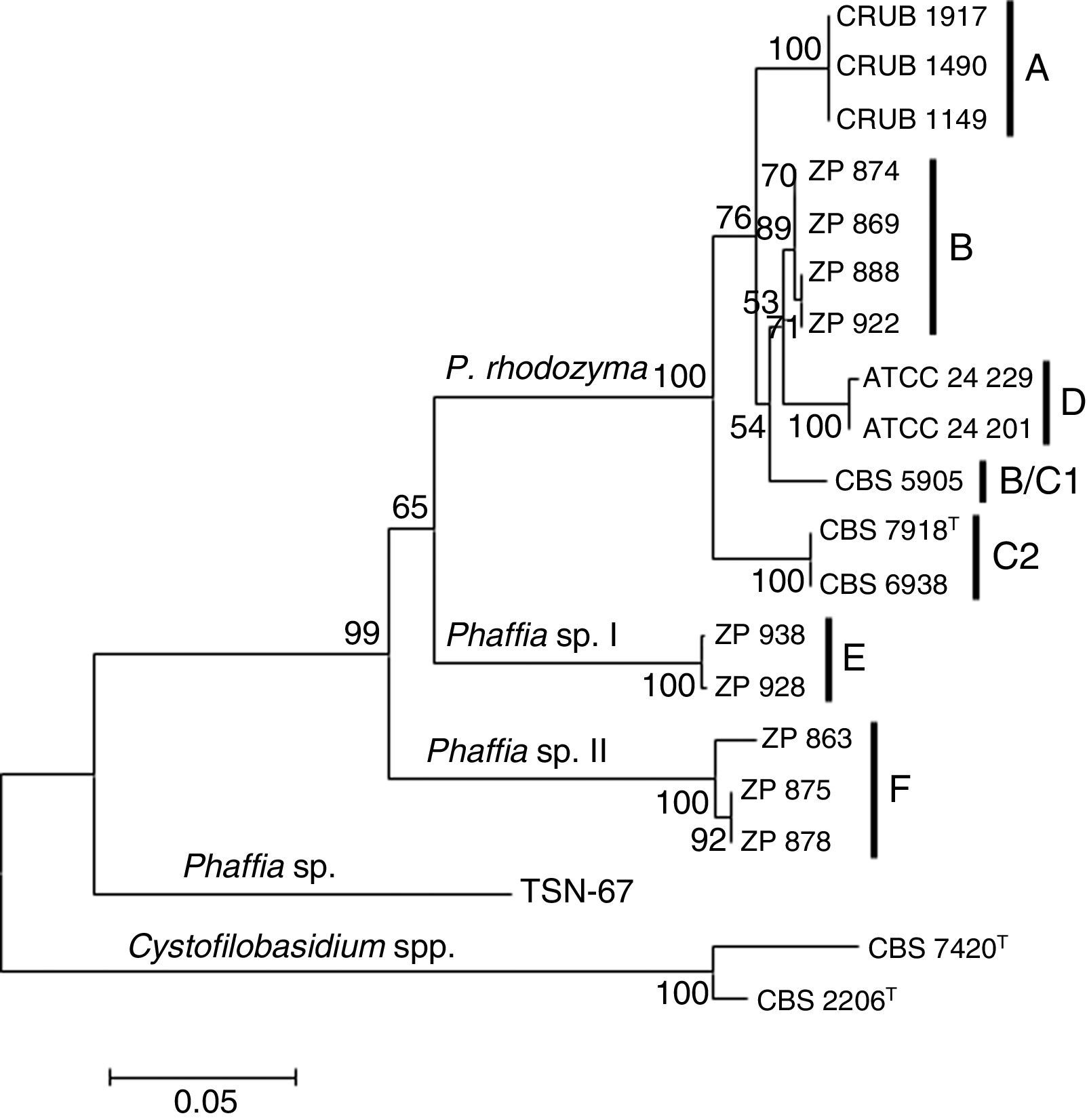
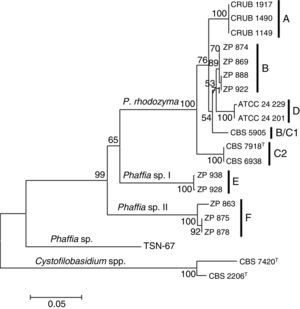
The Phaffia sp. strain TSN-6733 was negative for the specific primer. Two nucleotide differences were detected in the 3′ end of the specific primer for this strain explaining the lack of amplification (Table 3), which is in accordance with its relative distant phylogenetic relationship with the Phaffia clade (Fig. 2). Based on our phylogenetic analysis, the strain belongs to the order Cystofilobasidiales (Tremellomycetes, Agaricomycotina), occupying a basal position and having P. rhodozyma as the closest match, but showing more than fifty substitutions in the ITS region.
PhR primer specificity based on the nucleotide sequence of the annealing region
| Specie | Sequence | Number of mismatches |
|---|---|---|
| P. rhodozyma | 5′-GAC TTG TAC ACA GGC CGG CA-3′ | 0 |
| TSN-67 | 5′-GAC TTG TAC ACA GGC CGA CG-3′ | 2 |
| Cystofilobasidium spp.a | 5′-GAC TTG TAC ACA GGC CAA CG-3′ | 3 |
| Cystobasidium minutumb | 5′-GAC TCA TAC ACG GGC CGG CA-3′ | 3 |
| Dioszegia spp.c | 5′-GAC TTAGAC ACG GTC CGG CA-3′ | 4 |
Neighbor joining phylogenetic tree of Phaffia, based on internal transcribed spacer sequences. Bootstrap values (1000 replicates) and lineages are indicated. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The percentage of trees in which the associated taxa clustered together is shown next to the branches. The outgroup is constituted by Cystofilobasidium capitatum (CBS 7420) and Cystofilobasidium macerans (CBS 2206).
Carotenogenic species of the genus Cystofilobasidium were negative for the test since the specific band was absent due to the presence of 3 or more nucleotide substitutions mostly in the 3′ end. An environmental strain of Dioszegia spp., which was also tested, was negative (data not shown). In silico analysis of the remaining 5 Cystofilobasidium species, all Dioszegia known species and representative species of pigmented yeasts of the genus Rhodotorula, Rhodosporidium and Cystobasidium (ex-Rhodotorula) showed that at least 3 nucleotide differences were present in all cases (Table 3). Cystofilobasidium species were those showing the lower number of substitutions while Dioszegia species differed in 4 or 5 substitutions. With the exception of Cystobasidium minutum (Cystobasidiales) that had only 3 substitutions, the red yeasts of Rhodotorula and Rhodosporidium had 6 or more substitutions.
DiscussionBased on our results, the specific primer PhR, coupled with the universal primers ITS3 and NL4, has the required sensitivity and specificity for proper detection of relevant astaxanthin-accumulating yeasts of the genus Phaffia, here represented by all seven lineages recently described by David-Palma et al2. Despite the wide genetic diversity of this group of yeasts, the test detected all desired strains, differentiating them from other co-occurring, also carotenogenic and closely related yeast species, such as members of the genus Cystofilobasidium (Fig. 1).
The specificity of the method for astaxanthin producing strains is based on the complete homology of the PhR primer to the 5′ portion of the D1D2 domains of the large ribosomal subunit (26S), and the presence of 2 or more nucleotide differences in the 3′ end for distantly related strains (TSN-67) and Cystofilobasidium spp.
In a previous report, a new and innovative strategy for improving P. rhodozyma recovery rate in environmental samples was obtained26, which was successfully employed in the work of David-Palma et al2. A rapid identification method based on the simultaneous presence of astaxanthin and mycosporines was also described as a helpful tool for the screening of P. rhodozyma in a large set of new isolates. Although useful, this biochemical test is not as precise and reliable as DNA-based methods, hence we developed here a new molecular strategy (multiplex PCR reaction) for the accurate and rapid detection of astaxanthin-producing yeasts among environmental isolates or even for a rapid identity check of laboratory or production strains. In this regard, several successful developments with similar strategies for the molecular identification of Saccharomyces spp. and Zygosaccharomyces spp. have been reported7,19,21,23.
The use of three primers in a multiplex reaction has the advantage of still obtaining an amplicon in a negative sample, therefore, the results are unambiguous and cannot be attributable to a failure in the amplification reaction. The cost, complexity and time required for the test is low enough to be performed in most microbiology labs in a routine manner and could be even expanded to perform a colony PCR assay. Our own experience shows rRNA gene amplification (such as the one implied here) of Phaffia strains and other basidiomycetous yeasts can be easily achieved using the direct colony PCR method described by Espinar et al.3.
Ecological and biodiversity studies in new environments have led to the description of highly divergent astaxanthin-producing yeasts that were hitherto unknown. The biotechnological relevance of these new genetic lineages remains to be studied; however, the fact that other genetically different strains of Phaffia might be out there has been proved. Contreras et al.1 recently obtained novel strains of P. rhodozyma from Antarctic environments, one of which showed improved astaxanthin production with respect to the average yields of wild strains. The methods described earlier13,26 in combination with the molecular method presented here should represent excellent tools for the successful hunting of more astaxanthin-accumulating yeasts in the wild.
Ethical disclosuresProtection of human and animal subjectsThe authors declare that no experiments were performed on humans or animals for this investigation.
Confidentiality of dataThe authors declare that no patient data appears in this article.
Right to privacy and informed consentThe authors declare that no patient data appears in this article.
Conflict of interestThe authors declare that they have no conflicts of interest.
This work was supported by Consejo Nacional de Investigaciones Científicas y Técnicas (PIP424), Agencia Nacional de Promoción Científica y Técnica (PICT2007 N° 1745 and PICT2011 N° 1814) and Universidad Nacional del Comahue B171. We are thankful to A. Yurkov and J. Sampaio for providing valuable strains and information for this study.