Acidipropionibacterium acidipropionici is widely used for many applications, such as propionic acid production, cereal silage, and also as probiotic. Due to this plethora of applications, new isolates of A. acidipropionici with improved features are being searched for. These new isolates must be accurately identified, however, most approaches become expensive and time-consuming when the number of isolates is high. On the contrary, fluorescence in situ hybridization allows the affordable, reliable, and rapid identification of microorganisms in pure cultures and environmental and medical samples. Therefore, the aim of this work was to apply a fluorescent in situ hybridization probe for the reliable identification of new A. acidipropionici isolates. To this end, probe Pap446, specific for A. acidipropionici, was validated by hybridization assays with strains of this species from different origins, other species of the same genus or family, and unrelated genera. Eight isolates with propionibacterium characteristics were obtained from milk and feces of cows. Probe Pap446, hybridized only with isolates III and VI. The identity of these isolates was further confirmed by PCR using group and species-specific primers for propionibacteria and 16S rDNA sequencing.
Acidipropionibacterium acidipropionici es ampliamente usada para diversas aplicaciones, como producción de ácido propiónico, ensilado de cereales y probiótico. Debido a esta variedad de aplicaciones, continuamente se buscan nuevos aislamientos de A. acidipropionici con características nuevas. Estos nuevos aislamientos deben ser identificados correctamente, pero la mayoría de las técnicas disponibles resultan costosas e insumen mucho tiempo cuando el número de aislamientos es elevado. Por el contrario, la hibridación fluorescente in situ permite una identificación barata, confiable
y rápida de microorganismos en cultivos puros y en muestras ambientales y médicas. Por lo tanto, el objetivo de este trabajo fue la aplicación de una sonda oligonucleotídica en un protocolo de hibridación fluorescente in situ para la identificación confiable de nuevos aislamientos de A. acidipropionici. Con este fin, se validó la sonda Pap446, específica de A. acidipropionici mediante ensayos de hibridación con cepas de esta especie de diferente origen, otras especies del mismo género o familia, y géneros no relacionados. Se obtuvieron ocho aislamientos con características de propionibacterias a partir de leche y heces de vacas. La sonda Pap446, hibridó únicamente con los aislamientos III y VI. La identidad de estos aislamientos fue confirmada a través de PCR con cebadores específicos para propionibacterias y para A. acidipropionici, y mediante secuenciación del ADNr 16S.
Acidipropionibacterium acidipropionici is widely used for many applications, such as propionic acid and B12 vitamin production, and probiotics for animals3 and humans6. Due to this plethora of applications, new isolates of A. acidipropionici with better features are being searched for from various sources. Several techniques can be used for their accurate identification. Nevertheless, when the number of isolates to be identified is high, most approaches become expensive and time-consuming. On the contrary, the fluorescence in situ hybridization (FISH) technique allows the inexpensive, reliable, and rapid identification of microorganisms in pure cultures, environmental and medical samples. FISH is based on the hybridization of fluorescently labeled probes to complementary 16S rRNA sequences in permeabilized microbial cells, allowing their location and identification13. Therefore, the aim of this work was to apply a fluorescent in situ hybridization probe for the reliable identification of new A. acidipropionici isolates from milk and feces of cows
The presence of A. acidipropionici was investigated in milk and feces obtained from healthy Holstein dairy cows in a rural area of Tucumán (Argentina). For milk collection, the teat ends were disinfected with 70% ethanol and foremilk samples (n=10) were collected in sterile plastic tubes. Samples of cow manure (n=10) were aseptically transferred from the center of the dung into sterile plastic tubes. The samples were refrigerated and transported to the laboratory for immediate processing. The milk and manure samples were serially diluted in phosphate-buffered saline (130mmol/l NaCl, 10mmol/l sodium phosphate buffer, pH 7.4) (PBS) and plated onto modified Lactate agar [24ml/l sodium lactate (60% v/v), 30g/l casein peptone, 30g/l yeast extract, 125mM lithium chloride and 15g/l agar, pH 6.8]. The plates were incubated for ten days at 37°C in an anaerobic atmosphere provided by Anaerocult A (Merk, Germany) in an anaerobic jar (AnaeroGen system, Oxoid, UK). Convex and punctual colonies with creamy texture were transferred to LAPTg agar and incubated at 37°C for ten days in anaerobic conditions. Gram-positive cultures of short or filamentous rods in arrangements that resemble V, Y, or Chinese characters were selected and stored at −20°C in 10% (w/v) reconstituted non-fat milk supplemented with 15% glycerol. On the other hand, the following strains of dairy and intestinal origin belonging or not to the propionibacterium group were used as reference strains for FISH studies: A. acidipropionici LET 102, LET 103, LET 105, LET 107, LET 109 (intestinal origin), CRL1198, and ATCC 25562, A. jensenii CRL928, TL 219, TL 246, and TL 494, Propionibacterium freudenreichii CRL757, CRL758, TL 215, TL 253, TL 502, and TL 503, Streptococcus termophilus CRL 395 and CRL 396, Lactobacillus delbrueckii subsp. bulgaricus ATCC 11842, L. helveticus ATCC 15009 and ATCC 15807, and L. casei subsp. casei ATCC 393 (diary origin). All strains used as reference of A. acidipropionici species were previously identified by 16S rRNA gene sequencing, and their sequences are available in the database. All reference strains were stored in the same way as the selected isolates. Before use, the microorganisms used in this study were activated by three successive transfers in LAPTg broth incubated for 30h at 37°C.
Three species-specific oligonucleotide probes were used in this study for FISH: Pap44611, Pj4464, and Pfr4354. The specificity of these probes was confirmed both in silico, using the Probe match function of the Ribosomal Database Project, and in vitro, using the reference strains described above. Probes Eub338 and Non338 were used as positive controls of the permeability of fixed cells for rRNA-targeted oligonucleotides and as a negative control, respectively, as indicated by Babot et al.4. The 5′ end 6-FAM labeled probes were purchased from Sigma–Aldrich (Argentina). Designations and sequences of all probes are indicated in Table 1.
Sequence and bacteria targeting of oligonucleotide probes and primers used for FISH and PCR identification/16S RDA amplification, respectively.
| Probe | Sequence (5′→3′) | Target species | Temperature (°C) | |
|---|---|---|---|---|
| Ha | W | |||
| Pap446 | ACACCCCAAAACGATGCCTTCGCC | A. acidipropionici | 45 | 50 |
| Pj446 | CACCCCGATAGGCACTTCGTC | A. jensenii | 50 | 55 |
| Pfr435 | CTTGCGCTTCGTCATGGATGAAAG | P. freudenreichii | 45 | 50 |
| Eub338 | GCTGCCTCCCGTAGGAGT | Eubacteria | 45 | 48 |
| Non338 | ACTCCTACGGGAGGCAGC | None | 45 | 48 |
| Primer | ||||
| PB1 | AGTGGCGAAGGCGGTTCTCTGGA | Acidipropionibacterium, Propionibacteria, and Cutibacterium | – | – |
| PB2 | TGGGGTCGAGTTGCAGACCCCAAT | – | – | |
| PF | CTTTCATCCATGACGAAGCGCAAG | P. freudenreichii | – | – |
| PJ | GACGAAGTGCCTATCGGGGTG | A. jensenii | – | – |
| PA | GACGAAGGCATTCTTTTAGGGTGT | A. acidipropionici | – | – |
| 27F | GTGCTGCAGAGAGTTTGATCCTGGCTCAG | Universal | – | – |
| 1492R | CACGGATCCTACGGGTACCTTGTTACGACTT | Universal | – | – |
For bacterial cell fixation, 5ml of exponentially growing cultures were centrifuged (8000×g, 10min, 4°C), washed with 1ml of PBS, and resuspended in the original volume in the same solution. Cell suspensions were mixed 1:3 with cold paraformaldehyde solution (4% w/v in PBS) and incubated at 4°C for 16h. The cells were finally centrifuged (8000×g, 10min, 4°C), suspended in equal volumes of PBS and cold 96° ethanol, and stored at −20°C until use.
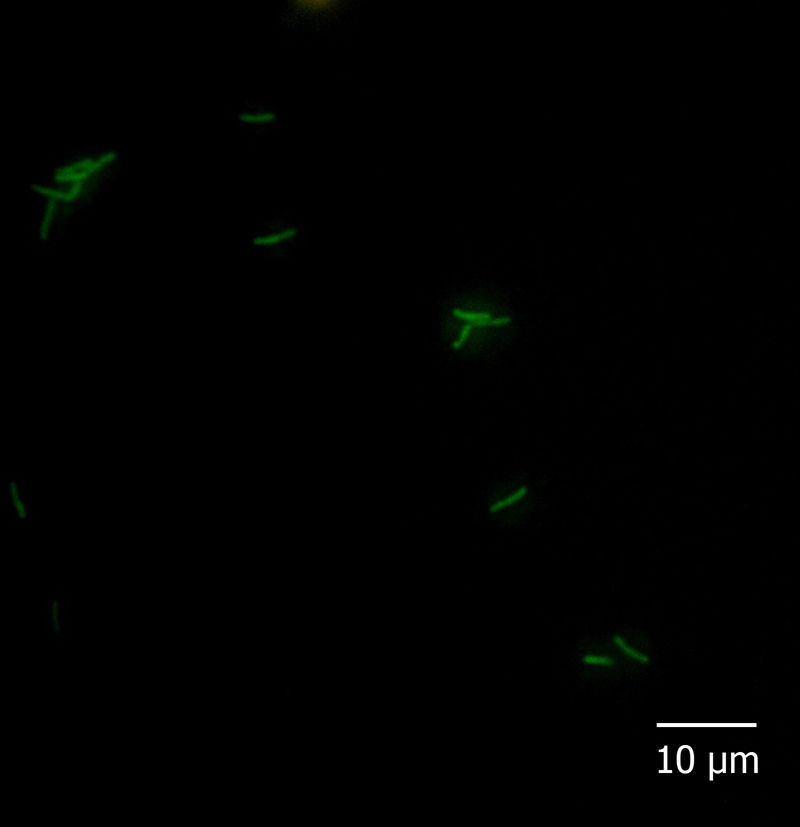
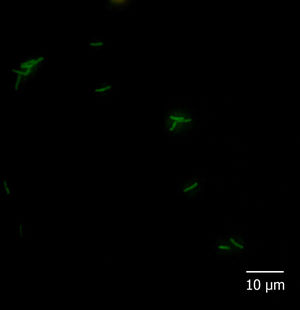
Five microliters of fixed cells were spotted onto glass slides precleaned in an ethanolic 10% (w/v) KOH solution and coated with a solution of 0.1% (w/v) gelatine and 0.01% (w/v) KCr(SO4)2 as described by Blasco et al5. Cells were dried at 37°C for 30min and finally dehydrated for 3min in each 50, 80, and 96% (w/v) ethanol solution, successively. Cell smears were covered with 1mg/l lysozyme (Sigma–Aldrich, Argentina) and incubated for 10min at 25°C. The enzymatic treatment was stopped by rinsing the slides thoroughly with water. The slides were dried and dehydrated by successive immersion for 3min in each 50, 80, and 96% ethanol solution. Then, hybridization buffer (20mmol/l Tris/HCl pH 7.2–7.4, 0.9mol/l NaCl, 0.01% SDS) containing 5ng/μl of probe solution was added to the cell smears on the microscopic slides and incubated overnight at 50 (probe Pj446) or 45°C (probes Pap446 and Pfr435) in a humid chamber. After hybridization, each slide was flooded with preheated washing buffer (20mmol/l Tris/HCl pH 7.2–7.4, 0.9mol/l NaCl) and incubated for 1h at a temperature 5°C higher than that used for hybridization. After rinsing in sterile distilled water, the slides were air-dried and stored in the dark. All the slides were covered with mounting medium (Inova Diagnostics Inc., USA) and coverslips, and observed at 1000× magnification with a fluorescence microscope Carl Zeiss Axio Scope A1 fitted with Filter Sets 01 (excitation: 365nm; emission: 397nm), 09 (excitation: 450–490nm; emission: 515nm; appropriated for 6-FAM), and 15 (excitation: 546nm; emission: 590nm). The absence of autofluorescence of bacteria in all filters, as well as the lack of fluorescence of hybridized cells in Filter Sets 01 and 15, was carefully checked.
To confirm the identity of isolates, DNA extraction for the propionibacterium group- and species-specific PCR was carried out according to Argañaraz-Martínez et al2. Primers PB1, PB2, PA, PJ, and PF12 were used for PCR assays (Table 1). The reaction mixture (20μl) consisted of 1.5mmol/l MgCl2, 2μl of 10× reaction buffer, a 100μmol/l concentration of each dNTP, a 0.5μmol/l concentration of each primer, 1μl bacterial DNA, and 1U of recombinant Taq DNA polymerase (Invitrogen, Argentina). The PCR reactions were performed in a MyCycler device (Bio-Rad Laboratories, USA), according to Rossi et al.12 with an annealing temperature of 60°C for the primer set PA-PB2 as the sole modification. The amplification products were separated by electrophoresis at 80V on 1.5% (w/v) agarose gel stained with SYBR® Safe DNA stain (Invitrogen, Argentina).
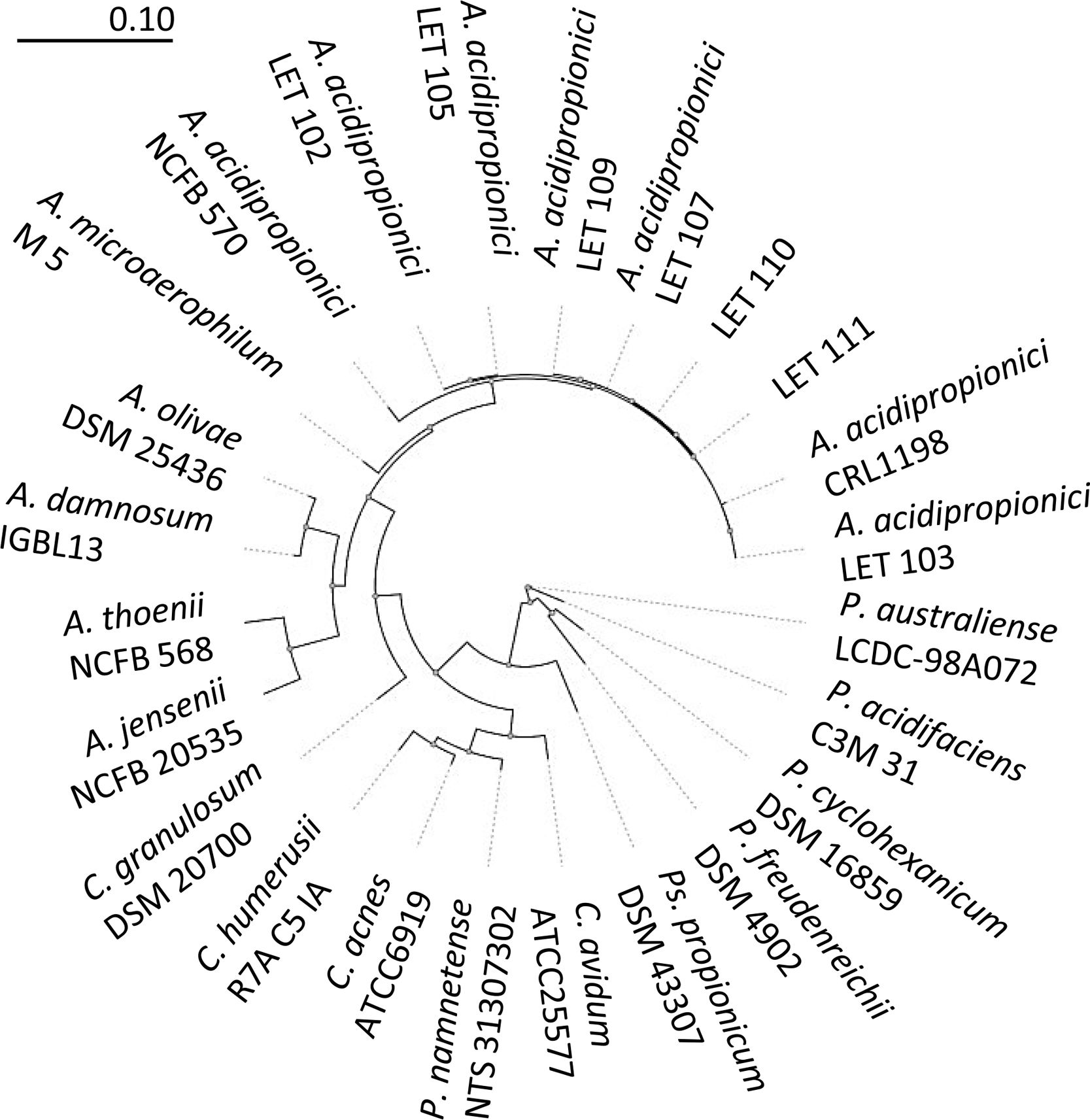
The amplification of the 16S rDNA of isolates presumptively identified as A. acidipropionici was carried out according to Argañaraz-Martínez et al.2 using primers 27F and 1492R (Table 1). The reaction mixture (50μl) consisted of 1.5mmol/l MgCl2, 5μl of 10× reaction buffer, 100μmol/l of each dNTP, 0.5μmol/l of each primer (27F and 1492R), 4μl bacterial DNA, and 1.5U of recombinant Taq DNA polymerase (Invitrogen, Argentina). The PCR reaction was performed in a MyCycler device (Bio-Rad Laboratories, USA). The amplification products were separated by electrophoresis at 80V on 0.8% (w/v) agarose stained with SYBR® Safe DNA Gel Stain (Invitrogen, Argentina), purified with 20% (w/v) polyethylene glycol (PEG8000, Sigma–Aldrich, Argentina) in 2.5mol/l NaCl solution, and sequenced at INTA-Castelar (Argentina) using a 3130xl Genetic Analyzer (Applied Biosystems, USA). The sequence fragments were assembled and edited using DNAMAN software (Version 4.03, Lynnon-Biosoft, Canada), and consensus sequences were compared to other 16S rDNA sequences in the EMBL/GenBank/DDBJ database using NCBI BLAST to determine their approximate phylogenetic affiliations. A phylogenetic tree was constructed using 16S rDNA sequences of type strains of Acidipropionibacterium and related genera with the tree builder function of PHYML9 using the maximum likelihood method with 500 bootstrap iterations.
Several culture media and strategies have been proposed for the selective isolation of members of the Propionibacterium group from diverse sources7. In our research group, the modified Lactate medium was successfully used for the isolation of A. acidipropionici and C. avidum strains from the intestinal content of chicks2. In the present study, eight convex and punctual colonies with a creamy texture resulting from independent samples were transferred from different modified Lactate agar plates to LAPTg agar plates. In this medium, the eight cultures thereby obtained were formed by Gram-positive short rods in arrangements that resemble V, Y, or Chinese characters. Three of these cultures (isolates I, II, and III) derived from milk, while the remaining cultures (isolates IV, V, VI, VII, and VIII) were obtained from feces. Propionibacteria are commonly isolated from cattle rumen and milk8 and they have been detected in cattle feces by meta-genomics10. However, to our knowledge, there are no reports of A. acidipropionici strains isolated from this niche.
With regard to the fluorescent in situ hybridization assay, all the reference strains and isolates hybridized with probe Eub338 and none of them with Non338. Moreover, among the species-specific probes, Pfr435 hybridized only with P. freudenreichii CRL758, CRL757, TL 215, TL 253, TL 502, and TL 503, Pj446 with A. jensenii CRL928, TL 219, TL 246, and TL 494, and Pap446 with A. acidipropionici CRL1198, ATCC 25562, LET 102, LET 103, LET 105, LET 107, and LET 109. These results showed that the hybridization was performed under adequate conditions and that probes Pap446, Pfr435, and Pj446 pair only with their targeted species. Besides Eub338, the isolates III and VI hybridized only with probe Pap446 (Fig. 1), which is specific for A. acidipropionici, while isolates I, II, IV, V, VII, and VIII failed to hybridize with all the species-specific probes used in this study. The FISH technique has been widely used for the in situ detection of different eukaryotic structures and microorganisms. However, this technique could also be applied for the rapid and affordable identification of pure cultures of bacteria, something especially useful when high numbers of isolates are studied1.
To confirm the identity of isolates III and VI as A. acidipropionici, DNA from all the isolates and one reference strain for each propionibacterium species was extracted and amplified with propionibacterium group- and species-specific primers. The primer set PB1-PB2 produced 610bp amplicons from DNA of the reference strains and isolates III and VI, indicating that these isolates belong to the propionibacterium group. With regard to species-specific primers, 867 and 864bp amplicons were obtained for P. freudenreichii CRL758 and A. jensenii CRL928 with primer set PF-PB2 and PJ-PB2, respectively, while the primer set PA-PB2 in combination with DNA from A. acidipropionici CRL1198 and isolates III and VI produced 868bp amplicons. The size of the amplification products agreed with that reported by Rossi et al.12 for A. acidipropionici. On the other hand, no amplification products were obtained with DNA from these isolates and the other primer sets studied. Moreover, DNA from isolates I, II, IV, V, VII, and VIII produced no amplification with neither of the primer sets used. Therefore, these isolates cannot be considered propionibacteria. Further studies are needed to identify these isolates, which showed remarkable features such as the ability to grow in the elective modified Lactate agar medium. Nevertheless, they exhibited growth kinetics (Supplementary Fig. 1), organic acid production (Supplementary Table 1), and carbohydrate fermentation patterns (Supplementary Table 2) clearly different from those of propionibacteria; therefore, these isolates could be ruled out as members of this group without the need of sequencing their 16S rDNA. Although useful for the phenotypic characterization of propionibacteria due to some distinctive features of these bacteria, these techniques are time-consuming and inconvenient when working with many isolates.
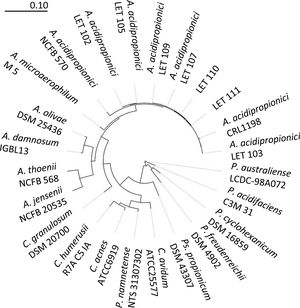
To further confirm the identity of isolates III and VI, PCR amplification and sequencing of their 16S rDNA was performed. Thus, 1300–1500bp nucleotide sequences were obtained for these isolates and deposited in the EMBL Nucleotide Sequence Database (http://www.ebi.ac.uk/embl/Submission/index.htmL) under accession numbers MW048631 (isolate III, renamed as A. acidipropionici LET 110) and MW048632 (isolate VI, renamed as A. acidipropionici LET 111). The query sequences of A. acidipropionici LET 110 and LET 111 showed the highest BLAST scores (2590-2518; E-value 0.0) and 99% maximum identity with A. acidipropionici strains. Furthermore, the phylogenetic tree constructed including most species of the propionibacterium group (Fig. 2) showed a close relationship between A. acidipropionici LET 110 and LET 111 and the reference strain A. acidipropionici NCFB 570.
Maximum likelihood tree of Acidipropionibacterium, Cutibacterium, Propionibacterium, and Pseudopropionibacterium species along with isolates A. acidipropionici LET 110 and LET 111. The tree was constructed using PHYML with 500 bootstrap iterations. Branch lengths are proportional to the number of substitutions per site (see scale bars).
In conclusion, the fluorescence in situ hybridization protocol reported in this work led to the reliable, rapid, and affordable identification of two isolates obtained from cattle milk and feces as A. acidipropionici. The reliability of probe Pap446 was supported by accessory assays such as PCR with species-specific primers and 16S rDNA sequencing. To our knowledge, this is the first report of the isolation of A. acidipropionici from cow feces.
FundingThis work was supported by Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) [grants number PIP 2015-0678 and PUE 2017-0035], Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT) [grants number PICT 2015-3714 and PICT 2016-0528], and Universidad Nacional de Tucumán (UNT) [grant number PIUNT D643/4].
Conflicts of interestThe authors declare that they have no conflicts of interest.