The surface of grapes lodges a complex community of yeast species responsible for spontaneous alcoholic fermentation. The study of indigenous Saccharomyces and “non-Saccharomyces” yeasts during grape must fermentation constitutes a major research area in microbial enology. Although there are detailed studies on the microbiota of Vitis vinifera L. grapes, little is known about the diversity of yeast communities present in non-vinifera Vitis ecosystems (i.e., grapes and spontaneously fermenting grape musts). Potentially scientific and/or enological valuable yeast strains from these non-vinifera Vitis ecosystems might never be isolated from V. vinifera L. In this updated review, we summarize relevant aspects of the microbial studies conducted on V. non-vinifera grapes and spontaneously fermenting grape musts.
La superficie de las uvas aloja una comunidad compleja de especies de levaduras responsables de la fermentación alcohólica espontánea. El estudio de estas levaduras Saccharomyces y «no-Saccharomyces» durante la fermentación del mosto de uvas constituye un área relevante de investigación microbiológica en enología. Si bien existen estudios detallados de la microbiota de uvas de Vitis vinifera L., poco se sabe sobre la diversidad de comunidades de levaduras presentes en ecosistemas de Vitis no-vinifera (i.e., uvas y mostos en fermentación espontánea). Cepas de levaduras presentes en ecosistemas de Vitis no-vinífera, con valor potencial científico y/o enológico, podrían no estar presentes en V. vinifera L. En esta revisión actualizada, resumimos los aspectos relevantes de los estudios microbiológicos efectuados en mostos en fermentación espontánea de uvas de V. no-vinifera.
Microbial communities present during grape must fermentation largely contribute to the sensory and organoleptic characteristics of wines4,16. Even in regular sulfur dioxide-treated must (i.e., used to limit and/or kill the endogenous microbiota and as a protective antioxidant agent), the inoculated yeast starters coexist during fermentation with surviving indigenous non-Saccharomyces and Saccharomyces yeast, fungi and bacteria species19, shaping the final sensory and organoleptic profile of the produced beverages. In the case of spontaneous fermentation (i.e., non-sulfited or mild sulfited musts), a challenging and risky winemaking process with potentially unpredictable outputs, the entire indigenous microbial community present in the must conducts the alcoholic fermentation11,19,29. Due to its scientific and industrial importance, the study of indigenous microbial communities in grapes and spontaneously fermenting musts is a major research area in enology19,29,42,43. Thus, different culture-dependent and/or metagenomic approaches, as well as DNA-based strategies, have been used to isolate and identify the complexity and population dynamics of microorganisms in enological ecosystems5,26,27.
Indigenous yeast diversity in V. non-vinifera ecosystemsMost of the studies on the microbiota of grapes and fermenting grape musts involve Vitis vinifera ecosystems3,12,29,39. Some of these studies suggest that the grape varieties themselves condition the microbial population structure during spontaneous fermentation8,13,24,36,39. Supporting this idea, vineyards cultivating different grape varieties appear to harbor more diverse Saccharomyces cerevisiae and non-Saccharomyces strains than vineyards cultivating only one grape variety13,36. In addition, it has been observed that particular yeast species show preferences for certain grape varieties (e.g., red or reddish basidiomycetes predominate in white grapes, while equal amounts of ascomycetes and basidiomycetes were observed on red grapes)32. Thus, specific structural and/or general physicochemical grape varietal factors appear to influence the structure and fitness of certain yeast microbiota24. Apparent specific associations between different Vitis and yeast species have recently been recognized34 (see below).
Non-Saccharomyces are the predominant yeasts isolated at the early stages of the spontaneous fermentation of V. vinifera grape musts12,19,29, with the most important genera being Hanseniaspora, Candida, Pichia and Metschnikowia19,42. By mid-fermentation, the population of non-Saccharomyces species decreases and the wine yeast S. cerevisiae completes the fermentation process1. A similar pattern of non-Saccharomyces and Saccharomyces yeast species succession was evidenced during the fermentation of V. non-vinifera grapes4,6,34. Baffi et al.4 identified Hanseniaspora uvarum as the most frequent non-Saccharomyces yeast species in the Isabel and Bordeaux varietals of Vitis labrusca grapes and must. Additionally, Issatchenkia occidentalis was the second and Issatchenkia orientalis the third most frequent yeast species isolated from Bordeaux grapes and Bordeaux/Isabel grapes and musts at all stages, respectively4. In a later study using Isabel and Bordeaux grapes from the same region6, H. uvarum was also a dominant yeast species, both on V. non-vinifera grape surfaces and at the initial stages of spontaneous fermentation. Pichia kluyveri was found at the beginning of fermentation while I. orientalis was isolated at the final stages of fermentation6. In both studies, S. cerevisiae was the most frequent yeast species during the middle and final phases of spontaneous fermentation4,6.
Important differences have been identified in the diversity and identity of non-Saccharomyces species isolated in V. vinifera and V. non-vinifera ecosystems. For example, two independent studies on the V. non-vinifera grape varieties Isabel and Bordeaux (V. labrusca), found a higher yeast diversity in the Bordeaux grapes than the Isabel grapes, suggesting that yeast diversity might be characteristic of each grape variety4,6. Similar yeast diversity was evidenced in the study of Danish grape varieties22. The hybrid variety ‘Leon Millot’ (V. vinifera and V. riparia×Vitis rupestris) reveals the same, or even higher, yeast diversity compared to the interspecific varieties (back crossings to V. vinifera) Rondo and Zalas Perle22.
Using a standard culture-dependent strategy, the population of non-Saccharomyces and Saccharomyces yeast species was recently studied on Isabella (V. labrusca L.) fermenting grape must in Argentina (i.e., vintage of year 2015)34. The dynamics of the yeast population during spontaneous fermentation of Isabella proved to be similar to that described for V. vinifera. Starmerella bacillaris, however, was the main yeast species at the early stages of spontaneous fermentation of Isabella must, dramatically decreasing its contribution in the middle and late stages of the process34. This dominance of S. bacillaris in the same ecosystem, however, was not observed in fermenting Isabella grapes from vintage of year 201733. Additionally, rare non-Saccharomyces yeast species were also recognized in Isabella must at the initial stages of fermentation, including Candida azymoides, Pichia cecembensis, Candida californica, Candida bentonensis, Issatchenkia hanoiensis and Candida apicola (Fig. 1A). Interestingly, some yeast genera commonly isolated from V. vinifera L. grapes and musts19 (e.g., Hanseniaspora, Torulaspora and Metschnikowia) were rarely identified and almost never dominated the yeast flora in the V. labrusca L. must analyzed34. These observations reinforce the research interest in biodiversity and extraordinary wine yeasts in ecological niches alternative to traditional V. vinifera ecosystems34.
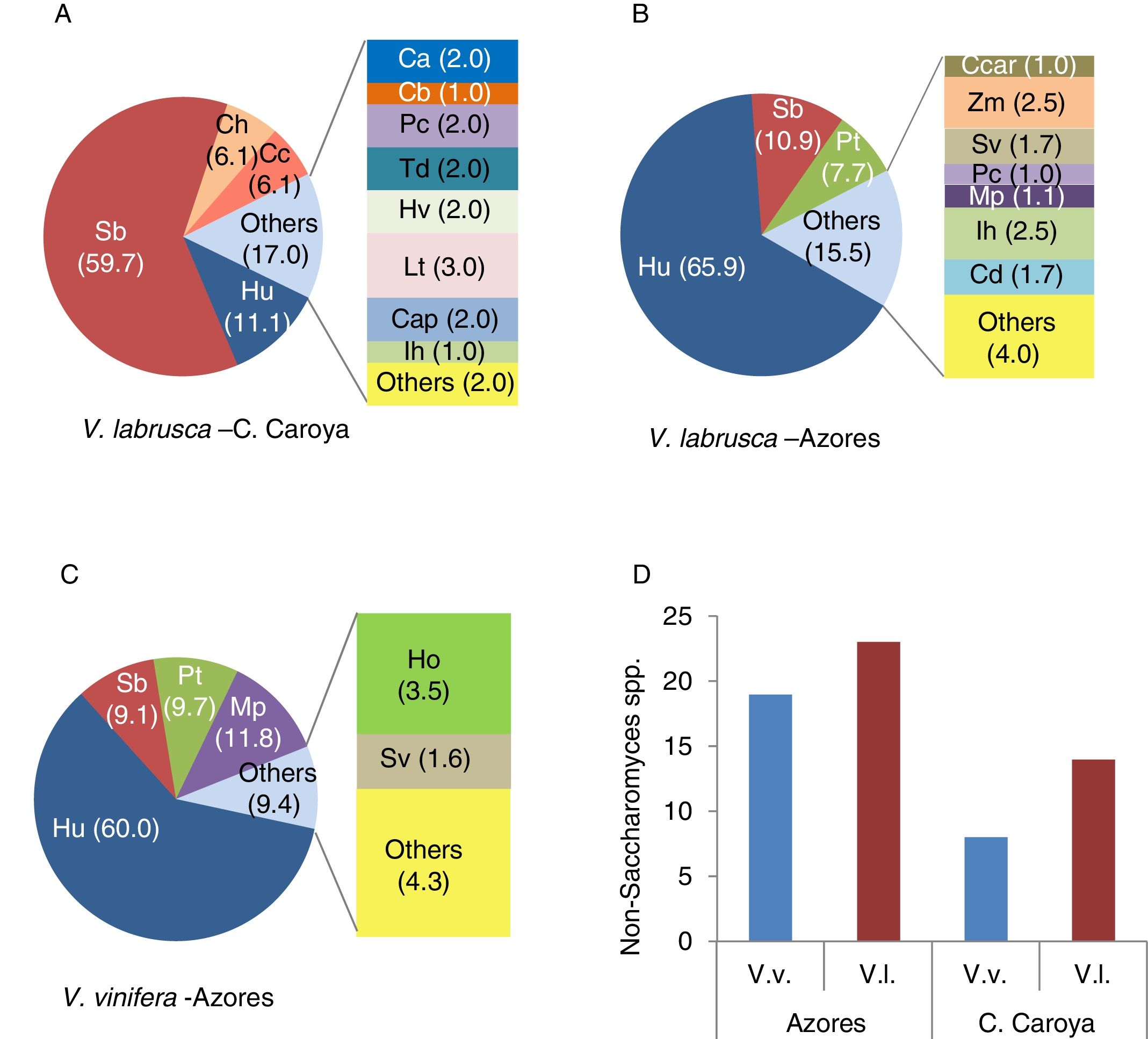
Diversity of non-Saccharomyces species isolated from grapes and spontaneously fermenting grape must from V. vinifera and V. labrusca vineyards from the Azores islands (Portugal) and C. Caroya (Córdoba, Argentina). (A, B, C) Relative contribution of non-Saccharomyces yeast species representing more than 1% of the isolates from spontaneously fermenting V. labrusca grape must from C. Caroya34 (A) and grapes from V. labrusca (B) and V. vinifera (C) from the Azores islands14,15. Numbers in parentheses indicate percentages. (D) Total number of non-Saccharomyces species identified on V. vinifera (blue bars) and V. labrusca (brown bars) grapes from the Azores islands (1710 and 3150 isolates, respectively) (data obtained from Refs. 14 and 15 and spontaneously fermenting grape musts from C. Caroya (40 and 100 isolates, respectively) (data obtained from Refs. 34 and 33). Non-Saccharomyces species are: Ca (Candida azymoides), Cap (Candida apicola), Cb (Candida bentonensis), Cc (Candida californica), Ccar (Candida carpophila), Cd (Candida diversa), Ho (Hanseniaspora opuntiae), Hu (Hanseniaspora uvarum), Hv (Hanseniaspora vineae), Ih (Issatchenkia hanoiensis), Lt (Lachancea thermotolerans), Mp (Metschnikowia pulcherrima), Pc (Pichia cecembensis), Pk (Pichia kluyveri), Pt (Pichia terricola), Sb (Starmerella bacillaris), Sv (Saccharomycopsis vini), Td (Torulaspora delbrueckii) and Zm (Zygoascus meyereae).
In the Azores Archipelago, different yeast microbiotas were identified on grapes harvested during vintages of years 2009 and 2010 from active versus abandoned V. labrusca vineyards14 (Fig. 1B) as well as on grapes from V. vinifera vineyards15 (Fig. 1C). In these studies, no apparent associations between grapevine and yeast species were found. Climatic conditions and geographic location seemed to be the underlying causes for the distribution of the predominant yeast species14. Interestingly, P. cecembensis and C. azymoides found on these V. labrusca grapes, two yeast species not previously recognized in either V. vinifera grapes or musts, were also found in the study on V. labrusca L. grapes in Argentina34 (Fig. 1A). These observations strongly suggest that at least these two yeast species are associated with V. labrusca L. grapes, regardless of their geographic origin and/or the associated human interventions. Moreover, in both locations I. hanoiensis, a yeast species rarely isolated in V. vinifera grapes, was also identified in V. labrusca grapes14,34. These results suggest that C. azymoides and P. cecembensis are preferentially associated with V. labrusca L. grapes and that specific Vitis-microbial interactions may underlie the assembly of specific grapevine yeast communities34. The great diversity of non-Saccharomyces species recognized in the V. labrusca and V. vinifera ecosystems studied by Drumonde-Neves et al.14,15 and Raymond Eder et al.34 is illustrated in Fig. 1.
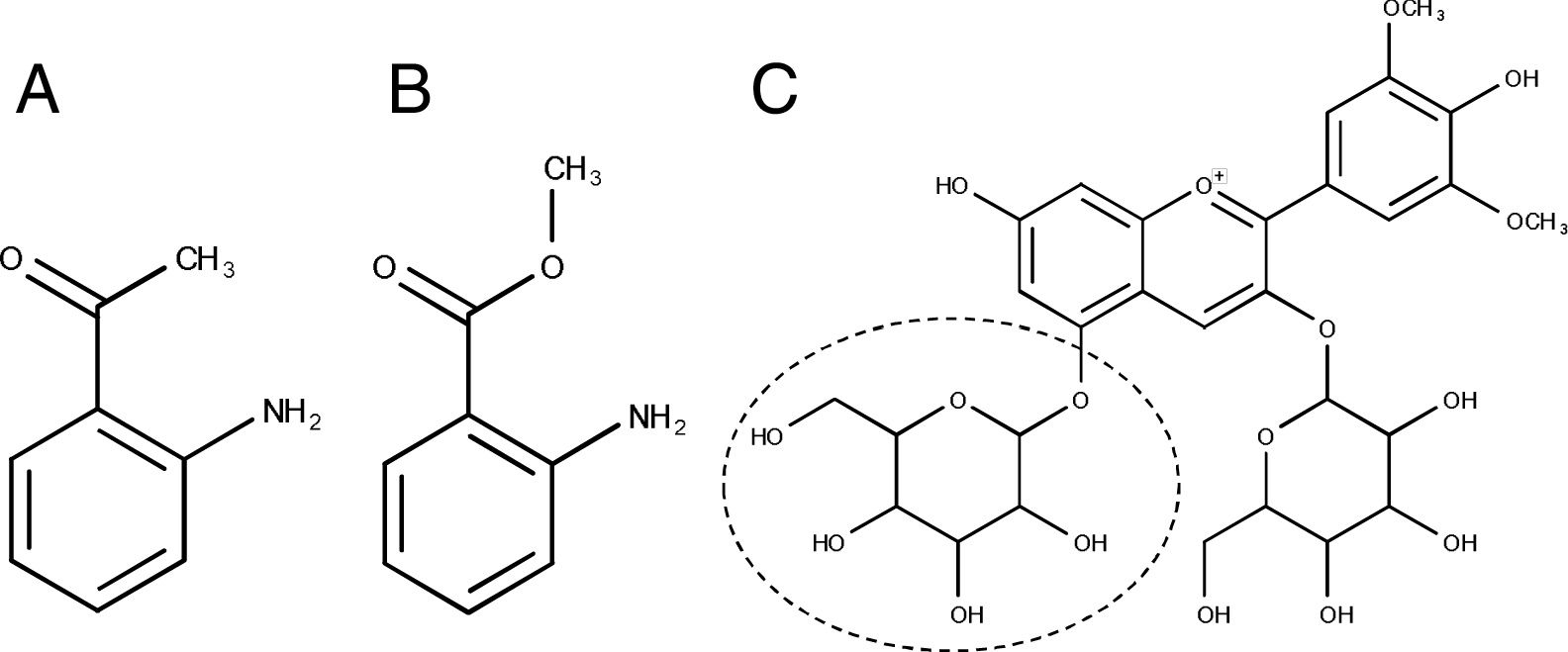
Microbial contributions to the sensory profiles of V. non-vinifera fermented beveragesV. non-vinifera species and their hybrids are popular in geographic areas where V. vinifera cannot develop properly10,18,23,45. Among these, V. aestivalis, V. labrusca, V. riparia and V. rotundifolia are widely used to produce wine, grape juice, table grapes and/or jam2,7,18,22,23,31. Remarkable sensory differences have been observed between wines obtained using V. vinifera and V. non-vinifera grapes7,41. Some V. non-vinifera fermented beverages are recognized as having disadvantages, including lower aroma complexity, high malic acid levels and/or increased amounts of some grape-derived vegetative odorants7,23. Five grape-derived “vegetal, earthy, minty” families of compounds are uniquely linked to some V. non-vinifera (i.e., V. riparia and V. cinerea) wines: eugenol (“clove”-like aroma), 1,8-cineole (also known as eucalyptol), cis-3-hexenol (“leafy-grassy” aroma), IBMP and IPMP (“herbaceous” and “earthy” aromas)41. Grapes from several American grape cultivars from V. labrusca (e.g., Catawba, Concord, Delaware, Isabella, Niagara, as well as some hybrids such as Agawam, Alexander and Onaka) are referred to as “foxy grapes” due to their intense fruity and/or artificial grape aroma/flavor notes in their wines30,41. The term “foxy” is used to describe a “unique, earthy and sweet muskiness” that can be perceived in these grapes. The presence of 2-aminoacetophenone and methyl anthranilate (Fig. 2A and B, respectively) is related to the perception of foxiness in V. labrusca grapes2,7,41,43.
In addition to these grape-derived compounds, specific yeast fermentation-derived products, such as volatile phenols, furans and esters, are responsible for some flavor differences between V. non-vinifera and V. vinifera wines35,44. Some disadvantages of V. non-vinifera wines have partially been remedied using alternative yeast and/or bacterial starters17,18,37,46. For example, an acidophilic I. orientalis strain, isolated from Korean Campbell Early grape pomace, has been shown to use malic acid efficiently as the sole carbon source37. In mixed fermentations with S. cerevisiae W-3 (industrial wine yeast), this I. orientalis strain efficiently degraded malic acid of Campbell Early grape must, without significantly influencing alcohol fermentation20. Additionally, an improvement in wine color was observed in these fermented mixed cultures compared to grape musts fermented with S. cerevisiae alone20 When the same I. orientalis yeast cells were immobilized on oriental oak charcoal and alginate, a 91.6% reduction of malic acid content was observed after 30h treatment of Campbell Early wine17. In these treatments, however, a decrease in the color of the wine was observed17. Interestingly, the use of an indigenous H. uvarum strain starter, isolated from spontaneously fermenting Campbell Early grape musts, also improved the sensory profile of Campbell Early wine18. It has been shown that grapes from V. non-vinifera cultivars normally do not reach high total reducing sugar levels, leading to fermented beverages with lower levels of alcohol18,35 than V. vinifera L. wines. Lower levels of ethanol (∼1% v/v) observed at the end of fermentation of Isabella (V. labrusca) grape musts, compared to the expected values based on the initial concentrations of total reducing sugars, have repeatedly been observed33,34.
In addition to alternative yeasts, dual starters of S. cerevisiae and Oenococcus oeni have been used to attempt to reduce the V. non-vinifera wine acidity of Campbell Early musts47. Although the use of commercial O. oeni starters for malolactic fermentation did not result in a significant change of the organic acid profiles, improvements were found in the sensory characteristics of the wines (i.e., higher levels of volatile compounds and an increased synthesis of esters and higher alcohols)47.
In a remarkable study, Son et al.38 studied wines obtained with grapes from four different V. non-vinifera cultivars in Korea (i.e., Muscat Bailey A –V. labrusca-, Campbell Early –V. labrusca B.-, Kyoho –. labrusca L.- and Merou –V. coignetiae-). As the same starter (i.e., S. bayanus) and fermentation conditions were used, this study highlighted the specific characteristics of each of the V. non-vinifera grape varieties analyzed. l-proline was noticed as an important metabolite for grape variety differentiation38, as it is relatively non-assimilable by yeast under anaerobic conditions40. Anthocyanin profiles, among polyphenols, have also been used for grape varietal differentiation9,21,25,35. In V. vinifera red cultivars, only cyanidin, delphinidin, petunidin, peonidin and malvidin 3-monoglucosides (Fig. 2C) occur along with the corresponding acetyl, p-coumaroyl and caffeoyl derivatives. In V. non-vinifera (i.e. V. labrusca, V. rotundifolia and their hybrid grapes), on the other hand, glycosylation of these compounds at both positions 3 and 5 is common25 and the presence of malvidin diglucoside (malvidin-3,5-diglucoside) (Fig. 2C) allows recognition of V. non-vinifera-derived wines. Wines produced with grapes from some V. non-vinifera species may contain high levels of this diglucoside, with 15mg/l being the maximum acceptable limit according to the international code of enological practices of the OIV (International Organization of Vine and Wine)28.
ConclusionsExtensive research has been conducted on the microbiological communities present in V. vinifera L. enological ecosystems as well as the sensory and organoleptic properties of V. vinifera L. wines. The few studies conducted on non-vinifera Vitis ecosystems, however, have identified several chemical, sensory and microbiological characteristics of these fermented beverages with potential interest in enology. Among these characteristics are a great diversity of non-Saccharomyces yeasts which may carry fermentation assets of winemaking importance. The apparent specific associations observed between different yeasts and Vitis species suggest that some yeast strains may be exclusive to non-vinifera Vitis ecosystems. Physical and chemical determinants may favor specific biological interactions between different species of grapes and yeasts, allowing the assembly of specific grapevine microbiotas. In addition to the biological interest of the spontaneous assembly of microbial communities on fruits and plants, the enological microbial ecosystems of V. non-vinifera may allow to recognize strains of yeasts or bacteria of interest in enology.
FundingThis work was supported by PICT-2014-313 from the FONCYT (Argentina) and SIV-2015 from the Catholic University of Córdoba. MLRE is supported by a fellowship from the Argentine National Research Council (CONICET) and ALR is a Principal Investigator of CONICET (Argentina).
Conflict of interestThe authors declare that they have no conflicts of interest.
We thank M. Bravetti for help with Fig. 2 and J. Heywood for English editing.