To evaluate the impact of polymorphisms of TNF-alpha (TNFA) and IL10 genes and their association with clinical phenotypes of psoriatic arthritis (PsA).
Materials and methodsThe study included 104 unrelated Venezuelan individuals, grouped into 52 patients with PsA, who fulfilled the CASPAR criteria, and 52 healthy individuals with no family history of psoriasis. The polymorphisms of the TNFA and IL10 genes were determined by Single Specific Primer-Polymerase Chain Reaction (SSP-PCR).
ResultsThe GA genotype and A allele of the TNFA-238G/A polymorphism appears to confer protection against the development of PsA (OR: 0.31, 95% CI: 0.92–1.05, p=0.02). The GA genotype of the TNFA-308G/A polymorphism is associated with a late onset age of PsA (GA=60±13.17 years vs. GG=43.55±14.29 years, p=0.002), and the GG genotype of the IL10 -1082A/G polymorphism with a longer time interval between the onset of psoriasis and the development of PsA (GG=27.4±24.11 years, GA=5.47±7.23 years, AA=7.86±8.51 years, p=0.001). The CC genotypes of IL10-819 C/T and IL10-592 C/A confer risk of damage to distal interphalangeal joints (OR: 4.79, p=0.026).
ConclusionsThe TNFA-238G/A polymorphism plays an important role in the development of PsA in mixed-race Venezuelans. Likewise, TNFA-308 G/A, IL10 -1082 A/G, -819C/T, -592C/A polymorphisms may modify the clinical expression of PsA.
Evaluar el impacto de los polimorfismos de los genes TNFA e IL10 y su asociación con el fenotipo clínico de la artritis psoriásica (APs).
Materiales y métodosSe incluyó a 104 individuos venezolanos, no relacionados, agrupados en 52 pacientes con APs, que reunieron los criterios CASPAR, y 52 individuos sanos, sin antecedentes familiares de psoriasis. Los polimorfismos de los genes TNFA e IL10 se determinaron por PCR-SSP.
ResultadosEl genotipo GA y alelo A del polimorfismo TNFA-238G/A parecen conferir protección contra el desarrollo de APs (OR: 0,31, IC del 95%: 0,92 -1,05, p: 0,02). El genotipo GA del polimorfismo TNFA-308G/A está asociado con una edad de inicio de APs tardía (GA=60±13,17 años vs. GG=43,55±14,29 años; p=0,002) y el genotipo GG del polimorfismo IL10-1082A/G con un intervalo mayor entre el inicio de la psoriasis y el desarrollo de la APs (GG=27,4±24,11 años, GA=5,47±7,23 años, AA=7,86±8,51 años, p=0,001). Los genotipos CC de IL10-819 C/T e IL10-592 C/A confieren riesgo de daño a las articulaciones interfalángicas distales (OR: 4,79, p=0,026).
ConclusionesEl polimorfismo TNFA-238G/A desempeña un papel importante en el desarrollo de la APs en mestizos venezolanos. Asimismo, los polimorfismos TNFA-308G/A, IL10-1082A/G, -819C/T y -592C/A pueden modificar la expresión clínica de la APs.
Psoriasis (Ps) is an inflammatory skin condition that affects 2% of the population and between 6% and 42% of these patients can develop joint inflammation, which is called psoriatic arthritis (PsA). PsA produces a very heterogeneous clinical affectation, being able to affect the peripheral joints, the spine and the entheses with different patterns and severity. Both Ps and PsA are multifactorial diseases, in which the interaction between genes and environment plays an important role in their development. Epidemiological studies have shown a strong familial aggregation for both diseases.1
Tumor necrosis factor alpha (TNF-α) is a key molecule in the inflammatory response and it has been demonstrated that it plays an important role in the genesis and maintenance of Ps and PsA. In the lesions of the skin and the synovial membrane of patients with Ps and PsA there is an increased expression of TNF-α and its blockade with biological drugs substantially improves both diseases.2 Two polymorphisms, located in the promoter region of the TNFA gene, affect the expression of TNF-α and they have been widely studied. In vitro studies have shown that the A allele of the TNFA-308G/A polymorphism is associated with an increased expression of TNF-α3 and the A allele of the TNFA-238G/A polymorphism is related to a decreased expression.4 It has been studied the influence of TNFA-308G/A and TNF-238 G/A polymorphisms on Ps and PsA in different populations, observing different and in some cases contradictory results, probably due to ethnic and methodological differences.5–12
Another important cytokine is interleukin-10 (IL-10), which has mainly anti-inflammatory functions.13 It has been seen a decrease of this cytokine in the serum and lesions of patients with Ps,14 although some studies have shown an increase in the synovial fluid and the synovial membrane of patients with PsA.15 There are many polymorphisms in the promoter region of the IL10 gene, with functional repercussions on the gene expression.16 Different studies of the association of variability of the IL10 gene with PsA have been carried out in different populations, observing diverse results.6,8,17
Considering that the cytokines IL-10 and TNF-α play an important role in PsA and that the variability of the genes that encode them can affect the expression thereof, we proposed to evaluate the impact of the polymorphisms of the TNFA and IL10 genes and their association with the clinical phenotype of PsA.
Materials and methodsThe study was conducted in 52 unrelated Venezuelan patients with PsA who met the CASPAR criteria.18 Patients under 18 years of age, those who had a relative with PsA participating in our study and patients with identified or suspected overlapping with collagen diseases were excluded. The patients were referred from different rheumatology clinics of the country, both public and private, within a period of 14 months.
The control group consisted of 52 unrelated healthy Venezuelan individuals, with no personal or family history of Ps or PsA.
Demographic data and other characteristics of the disease were previously collected, and this information was used to establish clinical subgroups, severity and damage caused by the disease. Likewise, the most frequent comorbidities in these patients were collected.
The patients signed an informed consent approved by the Bioethics Committee of the IVIC, in its session N° 1570, DIR-1067/1570/2016 0130/09, and of the University Hospital of Caracas (Hospital Clínico Universitario de Caracas-UCV), in its ordinary meeting N° 16, CBE N° 52/2014.
Genomic DNA extractionThe genomic DNA was extracted from peripheral blood mononuclear cells according to the protocol described by Bunce, in which chloroform is used as organic solvent.19
Genotyping of the TNFA geneThe TNFA-308G/A (rs1800629) variant was determined using the primers described by Verjans et al.,20 but using a protocol modified by the laboratory, which consisted in increasing the annealing temperature of the primers to 64°C, thus optimizing the specificity of the reaction. These SNPs of the promoter region were analyzed by polymerase chain reaction using sequence specific primers (PCR-SSP). The TNFA-238G/A (rs361525) variant was determined using the primers and the protocol previously described by Magalhaes et al.12 Two reaction mixtures were prepared for each polymorphism (-238 and -308), one for the ancestral or wild type allele (-238G and -308 G, respectively) and other for the mutated or infrequent allele (-238A and -308 A, respectively).
Genotyping of the IL10 geneThe IL10-1082A/G (rs1800896), -819C/T (rs1800871) and -592C/A (rs1800872) variants were determined using the primers and the protocol described by Kingkeow et al.21 These SNPs of the promoter region were analyzed by PCR-SSP. Two reaction mixtures were prepared for each polymorphism (-1082, -819 and -592), one for the ancestral or wild type allele (-1082G, -819C and -592C, respectively) and other for the mutated or infrequent allele (-1082A, -819 T and -592A, respectively).
Statistical analysisThe allelic and genotypic frequencies were calculated by direct counting. The Hardy–Weinberg (H–W) equilibrium was calculated through the chi-square test (χ2). The statistical significance of the differences in frequencies (alleles, genotypes, haplotypes) between the groups was estimated by the χ2 test using 2×2 contingency tables. The p-values were considered significant when p<0.05. The correlations between the variability of the genes studied and the different clinical variables were performed using the statistical package IBM SPSS Statistic 17, applying the Student's t-test for comparisons of only 2 groups and ANOVA when there were more than 2 groups. The determination of haplotypes of the polymorphisms of the promoter region of the TNF (-308, -238) and IL10 (-1082, -819, -592) genes was performed with the SNPStats program.22
ResultsClinical and demographic characteristicsThe average age of the patients with PsA is significantly higher than that of the control group (54.42 years vs. 44.63 years, respectively; p=0.001); however, regarding gender, no significant differences were observed between both groups. Concerning the clinical characteristics, 48% of the patients had type 1 Ps, that is, the skin lesions appeared for the first time before the age of 40, and 38.5% had a family history of Ps. The joint disease was classified into 2 groups: a peripheral form that comprised 67.3% of patients and another form that in addition had axial involvement (sacroiliitis), called spondyloarthritis, which comprised 32.7% of patients. Other manifestations, such as affectation of entheses, nail involvement, dactylitis, radiological commitment, and the types of pharmacological treatments, as well as the composite indices of disease activity and function, such as DAS 28, BASDAI and HAQ, are shown in Table 1.
Clinical characteristics of the patients.
| Characteristic | Patients with PsA (n=52) |
|---|---|
| Type of psoriasis | |
| Type 1 | 48.1 (25) |
| Type 2 | 51.9 (27) |
| Age at onset of Ps | |
| Mean±SD | 39.11±13.37 years |
| Age at onset of PsA (mean±SD) | 46.7±15.42 years |
| Ps-PsA interval (mean±SD) | 9.3±12.57 years |
| PsA without Ps | 15.4 (8) |
| Relatives with psoriasis | |
| Yes | 38.5 (20) |
| No | 61.5 (32) |
| PsA pattern | |
| Peripheral | 67.3 (35) |
| Spondyloarthritis | 32.7 (17) |
| Enthesitis | 65.4 (34) |
| Dactylitis | 44.2 (23) |
| Nail involvement | 40.4 (21) |
| Sacroiliitis | 32.7 (17) |
| HLA-B27 positive | 5.8 (3) |
| DIP involvement | 25 (13) |
| RX erosions | 44.2 (23) |
| RX hyperproliferation | 7.7 (4) |
| DAS 28 (mean±SD) | 4.8±1.91 points |
| BASDAI (mean±SD) | 4.0±2.07 points |
| HAQ (mean±SD) | 0.6±0.69 points |
| Time to Dx of PsA (mean±SD) | 7.7±9.12 years |
| Metabolic disease | 21.2 (11) |
| TT only synthetic DMARDs | 42.3 (22) |
| TT only biological agents | 19.2 (10) |
| TT DMARDs synt+biol | 34.6 (18) |
| Only NSAIDs | 1.9 (1) |
The frequencies are shown in percentage and in parentheses the number of individuals.
NSAIDs: non-steroidal anti-inflammatory drugs; SD: standard deviation; DMARDs: disease modifying antirheumatic drugs; Dx: diagnosis; DIP: distal interphalangeal; RX: radiology; TT: treatment; synt+biol: synthetic and biological drugs administered together.
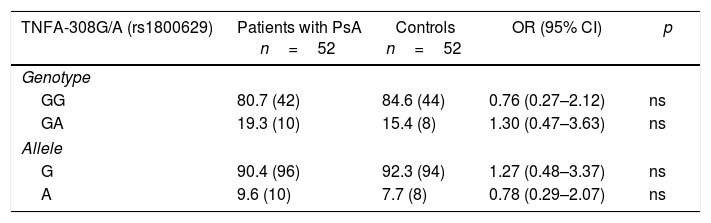
The existence of H-W equilibrium for the genotypic distribution of the polymorphic sites TNFA-308G/A and TNFA-238G/A was confirmed in healthy individuals. Table 2 shows the distribution of the allelic and genotypic frequencies of the TNFA-308G/A and TNFA-238G/A variants, observing for both polymorphisms and groups studied, only 2 of the 3 possible genotypes (GG and GA). When establishing comparisons of the genotypic and allelic frequencies of the TNFA-308G/A variant between patients with PsA and controls, no significant differences were observed. In contrast, for the TNF-238G/A variant, significantly increased frequencies were observed for the GG genotype (OR=3.22, 95% CI, 0.95–10.80, p=0.025) and the G allele (OR=2.95, 95% CI, 0.91–9.61, p=0.03) in the patients with PsA with respect to the controls, and a significantly increased frequency of the GA genotype (OR=0.31, 95% CI, 0.09–1.05, p=0.025) and the A allele (OR=0.03, 95% CI, 0.10–1.09, p=0.036) in the controls with respect to the patients.
Distribution of the genotypic and allelic frequencies of the -308G/A (rs1800629) and -238G/A (rs361525) variants of the TNFA gene in patients with psoriatic arthritis and healthy controls.
| TNFA-308G/A (rs1800629) | Patients with PsA n=52 | Controls n=52 | OR (95% CI) | p |
|---|---|---|---|---|
| Genotype | ||||
| GG | 80.7 (42) | 84.6 (44) | 0.76 (0.27–2.12) | ns |
| GA | 19.3 (10) | 15.4 (8) | 1.30 (0.47–3.63) | ns |
| Allele | ||||
| G | 90.4 (96) | 92.3 (94) | 1.27 (0.48–3.37) | ns |
| A | 9.6 (10) | 7.7 (8) | 0.78 (0.29–2.07) | ns |
| TNFA-238G/A (rs361525) | Patients with PsA n=52 | Controls n=52 | OR (95% CI) | p |
|---|---|---|---|---|
| Genotype | ||||
| GG | 92.3 (48) | 78.8 (41) | 3.22 (0.95–10.88) | 0.025 |
| GA | 7.7 (4) | 21.2 (11) | 0.31 (0.09–1.05) | 0.025 |
| Allele | ||||
| G | 96.2 (100) | 89.4 (93) | 2.95 (0.91–9.61) | 0.03 |
| A | 3.8 (4) | 10.6 (11) | 0.39 (0.10–1.09) | 0.03 |
The frequency is expressed as a percentage. The values shown in parentheses represent the number of individuals who are carriers of the genotype for the studied polymorphic site.
CI: confidence interval; ns: not significant; p: probability value (p<0.05=significant value).
In both studied groups was observed the existence of the 4 possible TNF-308/-238 haplotypes, which in order of frequency were: GG, GA, AG and AA. The latter was observed in an individual of the control group and is considered a rare haplotype due to the low frequency of the A allele in both TNFA polymorphisms. Likewise, no association was observed between haplotypes and PsA.
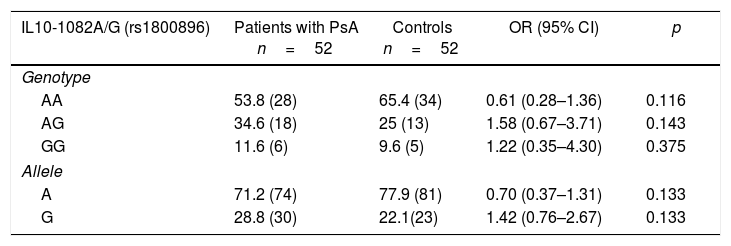
Distribution of the genotypic and allelic frequencies of the -1082 (rs1800896), -819 (rs1800871) and -592 (rs1800872) polymorphisms of the IL10 gene in patients with psoriatic arthritis and healthy controlsIt could be proven the existence of H-W equilibrium for the phenotypic distribution of the polymorphic sites IL10-1082A/G, IL10-819C/T and IL10-592C/A in healthy individuals. Table 3 shows the distribution of the allelic and genotypic frequencies of IL10-1082A/G, IL10-819C/T and IL10-592C/A variants, observing for each polymorphism the 3 possible genotypes in both studied groups. When establishing comparisons of the genotypic and allelic frequencies of the IL10-1082A/G, IL10-819C/T and IL10-592C/A variants between patients with PsA and healthy controls, no significant differences were observed.
Distribution of the genotypic and allelic frequencies of the -1082 (rs1800896), -819 (rs1800871) and -592 (rs1800872) variants of the IL10 gene in patients with psoriatic arthritis and healthy controls.
| IL10-1082A/G (rs1800896) | Patients with PsA n=52 | Controls n=52 | OR (95% CI) | p |
|---|---|---|---|---|
| Genotype | ||||
| AA | 53.8 (28) | 65.4 (34) | 0.61 (0.28–1.36) | 0.116 |
| AG | 34.6 (18) | 25 (13) | 1.58 (0.67–3.71) | 0.143 |
| GG | 11.6 (6) | 9.6 (5) | 1.22 (0.35–4.30) | 0.375 |
| Allele | ||||
| A | 71.2 (74) | 77.9 (81) | 0.70 (0.37–1.31) | 0.133 |
| G | 28.8 (30) | 22.1(23) | 1.42 (0.76–2.67) | 0.133 |
| IL10-819C/T (rs1800871) | Patients with PsA n=52 | Controls n=52 | OR (95% CI) | p |
|---|---|---|---|---|
| Genotype | ||||
| CC | 50 (26) | 42.3 (22) | 1.36 (0.62–2.95) | 0.216 |
| CT | 38.5 (20) | 36.5 (19) | 1.08 (0.49–2.40) | 0.420 |
| TT | 11.5 (6) | 21.2 (11) | 0.48 (0.16–1.43) | 0.093 |
| Allele | ||||
| C | 69.2 (72) | 60.6 (63) | 1.46 (0.82–2.59) | 0.096 |
| T | 30.8 (32) | 39.4 (41) | 0.68 (0.38–1.21) | 0.096 |
| IL10-592 (rs1800872) | Patients with PsA n=52 | Controls n=52 | OR (95% CI) | p |
|---|---|---|---|---|
| Genotype | ||||
| AA | 11.5 (6) | 21.2 (11) | 0.48 (0.16–1.43) | 0.093 |
| AC | 38.5 (20) | 36.5 (19) | 1.08 (0.49–2.40) | 0.420 |
| CC | 50 (26) | 42.3 (22) | 1.36 (0.62–2.95) | 0.217 |
| Allele | ||||
| A | 30.8 (32) | 39.4 (41) | 0.68 (0.38–1.21) | 0.092 |
| C | 69.2 (72) | 60.6 (63) | 1.46 (0.82–2.59) | 0.096 |
The values shown in parentheses represent the number of individuals who are carriers of the genotype for the studied polymorphic site. The frequency is expressed as a percentage.
CI: confidence interval; p: probability value (p<0.05=significant value).
In both groups studied it was observed the existence of 3 of the 4 possible haplotypes -IL10-1082/-592/-819, which in order of frequency were: ATA, ACC and GCC. However, no association was observed between the haplotypes and PsA.
Correlation between the clinical characteristics of patients with psoriatic arthritis and the polymorphisms of the TNFA and IL10 genesWhen correlating the different clinical manifestations with the genetic polymorphisms studied, differences in means and significant associations were observed between:
- 1.
The GA genotype of the TNFA-308G/A polymorphism and a later onset age to develop PsA (GA=60±13.17 years; GG=43.55±14.29; p=0.002).
- 2.
The GG genotype of the IL10-1082A/G polymorphism and a longer Ps- PsA interval of latency, i.e. more time between the appearance of the PsA after the onset of the Ps (GG=27.4±24.11 years, GA=5.47±7.23 years, AA=7.8±8.51 years, p=0.001).
- 3.
The presence of the T (TT or CT) allele of the IL10-819C/T polymorphism and the A (AA or CA) allele of the IL10-592C/A polymorphism with protection against erosions and loss of the joint space of distal interphalangeal joints (DIP) (OR: 0.20, 95% CI, 0.049–0.880, p=0.026). In contrast, the CC genotypes of -819C/T and -592C/A confer risk of damage to the DIP joints (OR: 4.79, 95% CI, 1.13–20.20, p=0.026).
No significant differences were observed for the other characteristics and manifestations of the disease.
DiscussionPsA is a multifactorial disease, where the interaction of a combination of variants or alleles common in the population with low penetrance, along with environmental triggering factors, determines the appearance of the disease.1 The clinical heterogeneity of PsA could be a reflection of the genetic heterogeneity that exists in these patients and some polymorphic genes involved in certain inflammatory pathways could be related to the risk of developing PsA or could be modifiers of the disease phenotype.1,23 Some cytokines have a considerable importance in the pathogenesis of the disease, and therefore it was proposed to study the polymorphisms of the genes that encode for 2 of these cytokines, such as TNF-α and IL-10. Although studies of the association between these genes and PsA have been conducted in European5–10 and Asian14,17,24 populations, they have not yet been described in the Hispanic-American mixed-race population, being this a first study of this type of population, in which differences with the results described in other populations were observed.
An association between the A allele and the GA genotype of the TNFA-238G/A polymorphism and decreased risk of developing PsA was observed in this study. In previous studies, carried out in Ps and PsA, it has been described a positive association between the A allele of TNFA-238G/A and the susceptibility to develop these 2 diseases.25,26 However, this association has been described mainly in European populations, but not in Asian populations.11,25 In several studies of Ps and PsA it has been demonstrated that in European populations, the A allele of the TNFA-238G/A polymorphism is in linkage disequilibrium with the HLA-Cw6 allele.5–7 This linkage disequilibrium is relevant in the risk association between PsA and TNFA-238G/A polymorphism because the HLA-Cw6 allele has been associated with type 1 Ps26 and PsA in patients in whom the skin lesions begin before age 40.27 Given that in European populations the A allele of TNFA-238 and HLA-Cw6 is in linkage disequilibrium, it could be considered that the A allele of the TNFA-238G/A polymorphism is not the causative variant, but rather the HLA-Cw6 allele could be responsible for this risk association.
One weakness of this study was the impossibility of typifying the HLA-Cw6 allele in all patients; however, in a study conducted in Venezuelan mixed-race individuals with Ps without PsA it was described a frequency of the HLA-Cw6 allele of 5.4% in healthy individuals with respect to a frequency of 21.8% in patients with Ps28; this prevalence of HLA-Cw6 is much lower than the one described in other populations, such as the European. Therefore, the association observed between the A allele and the GA genotype of the TNFA-238G/A polymorphism and protection against the development of PsA could be explained because the A allele of the TNFA-238G/A polymorphism has been associated with a decreased expression of TNF-α.4
An important difference between this study and other cohorts is that it was included a higher percentage of patients with Ps type 2; the latter could explain the difference between the results obtained and those published in other populations, since it has been demonstrated that these 2 groups of patients with Ps behave genetically differently, for example, the frequency of the HLA-Cw6 allele is not higher in patients with type 2 Ps than in the general population.26
Likewise, it was observed a significant association between the GA genotype of the TNFA-308G/A polymorphism and the late onset of the PsA. These results are consistent with a study carried out in Caucasians, in which the association of the GA genotype of the TNFA-308G/A polymorphism and an increased average age of onset of Ps is described7 and with a study conducted in Spanish patients with PsA, in which an increased frequency of the A allele of the TNFA-308G/A is observed in female patients with type 2 Ps.27
However, it is important to note that TNFA-857C/T polymorphism, located in the promoter region of the TNFA gene, has been associated with susceptibility to PsA in different European populations.29,30 As well, the rs12044149 variant, close to the IL23R gene, and the rs9321623 variant, close to the TNFAIP3 gene, have been strongly associated with PsA, but not with cutaneous Ps.31
When correlating the polymorphisms of the IL10 gene with the expression or phenotype of the disease (clinical variables of the disease), only 2 of them were significant. One was the interval or latency between the appearance of Ps and PsA, which was longer in those patients who presented the GG genotype of the IL10-1082A/G polymorphism. This association is interesting because this polymorphism is related to an increased production of IL-10, indicating that a higher concentration of this cytokine could be delaying the joint manifestations in those patients with a diagnosis of Ps, although it is important to mention that not all patients with Ps will develop PsA and that in not all patients with PsA, the skin lesions precede joint manifestations.32 However, it is still an interesting result to be confirmed with other methodologies, since it could have a therapeutic implication, for example, the use of recombinant IL-10 could prevent the appearance of joint manifestations in those patients who have a diagnosis of Ps.
The other clinical manifestation that resulted significantly associated with these IL10 polymorphisms was the commitment of the DIP joints, observing an association of the CC genotypes of IL10-819C/T and IL10-592C/A, respectively, and damage of the DIP joints. These genotypes have been associated with an increased production of IL-10,16 but is not easy to explain their association with this subgroup of patients with PsA. However, it has been described the presence of an increased subpopulation of CD163+ macrophages polarized to an M2 phenotype, in the presence of IL-10, in the synovial membrane of patients with PsA,33,34 although the role of these macrophages in the pathogenesis of PsA is still unknown. Another possible explanation is that the alleles of the IL10-819C/T and IL10-592C/A polymorphisms are in linkage disequilibrium with other nearby genes, which by other mechanisms, for example immune response or bone repair, are associated with DIP joint involvement. It is important to highlight that in some of the patients older than 40 years included in this study, it was not possible to differentiate whether the damage of these joints had occurred due to PsA or to osteoarthritis of the hands. However, in either of the 2 circumstances there are no previous works documenting this interesting association.
In conclusion, the results obtained allow us to consider that there is a risk association between TNFA-238G/A polymorphism and PsA in Venezuelan mixed-race patients. In contrast, the TNFA-308G/A and IL10-1082A/G, IL10-819C/T and IL10-592C/A polymorphisms modify the expression of the disease, such as the later onset of the PsA and the appearance of one of its manifestations, like the affectation of the DIP joints. This confirms the influence of genetic factors on PsA and indirectly demonstrates the relationship that exists between some cytokine pathways and the onset and development of this disease. Finally, studies should be conducted in a larger group of patients and in other Hispanic-American mixed-race populations in order to validate these findings.
FundingThis work was financed with funds of the Venezuelan Institute for Scientific Research.
Conflict of interestDr. Luis. Gutiérrez has a research grant (IIR/PFIZER).
Venezuelan Group for the Study of Spondyloarthritis (GRUVES).
Please cite this article as: Herrera F, Gutiérrez L, Salazar Alcalá E, Balbas O, Fernández Mestre M. Papel de los genes TNFA e IL10 en el desarrollo y manifestaciones clínicas de la artritis psoriásica. Rev Colomb Reumatol. 2018;25:9–15.