Candida dubliniensis is a germ tube and chlamydoconidia producing Candida species that may be misidentified as Candida albicans. Molecular-based methods are the most reliable techniques for C. albicans and C. dubliniensis differentiation. However, accurate, quick and inexpensive phenotypic tests are needed to be used in low-complexity mycology laboratories.
AimsTo evaluate colony morphotypes on Sabouraud-triphenyltetrazolium agar as a tool for C. dubliniensis and C. albicans differentiation.
MethodsThe morphology of 126 C. albicans and C. dubliniensis strains was evaluated and compared with their identification by molecular methods.
ResultsThe method showed 100% sensitivity and specificity when color and the presence or absence of large white mycelial halo was evaluated.
ConclusionsColony morphotype on Sabouraud-triphenyltetrazolium agar should be considered as a new tool to differentiate C. dubliniensis and C. albicans.
Candida dubliniensis es una especie del género Candida capaz de producir tubos germinativos y clamidoconidios, y puede ser identificada erróneamente como Candida albicans. Las técnicas moleculares de identificación son consideradas las más específicas para diferenciar estas especies. Sin embargo, se siguen necesitando métodos exactos, rápidos y de bajo coste para ser utilizados en laboratorios de micología de baja complejidad.
ObjetivosEvaluar el morfotipo de las colonias de levaduras en agar Sabouraud-trifeniltetrazolio como una herramienta para diferenciar C. dubliniensis de C. albicans.
MétodosSe evaluó la morfología de 126 aislamientos de C. albicans y C. dubliniensis y los resultados fueron comparados con los obtenidos utilizando métodos moleculares.
ResultadosEl método utilizado mostró una sensibilidad y una especificidad del 100% cuando se evaluó el color y la presencia o ausencia de un gran halo de micelio blanco.
ConclusionesLa evaluación del morfotipo de las colonias en agar Sabouraud-trifeniltetrazolio puede ser utilizada como una nueva herramienta para diferenciar C. dubliniensis de C. albicans.
Candida dubliniensis is a germ tube and chlamydoconidia producing Candida species described in 199515,17 that may be misidentified as C. albicans.7 Firstly, C. dubliniensis was associated with oropharyngeal candidiasis in HIV+ population15,17 but later it was also isolated from HIV negative patients and from different body sites and fluids, including blood, urine, etc.3,8,10 Molecular-based methods are the most reliable techniques for C. albicans and C. dubliniensis differentiation.2,12,16 However, accurate, quick and inexpensive phenotypic tests are needed to be used in low-complexity mycology laboratories. Different tests were proposed and have been used as screening methods, including differences in carbohydrate assimilation, growth capacity at 42°C, 45°C and in hypertonic media, clustered chlamydoconidia production, etc.1,4–6,9,13,14,16 Any of the described methods are 100% specific for C. dubliniensis and C. albicans differentiation. In this work, colony morphotype on Sabouraud-triphenyltetrazolium agar (STA) was evaluated as a tool for C. dubliniensis and C. albicans differentiation.
Colony morphotype data were obtained from 110 C. albicans and 12 C. dubliniensis clinical strains isolated from different sources (blood, urine, vulvovaginal infections, etc.). C. albicans ATCC 90028, C. albicans ATCC 36082, C. albicans Sc5314 and C. dubliniensis NCPF 3949 were included in the study as control strains. All the strains were identified by phenotypic5 and molecular methods.3,12 The molecular identification was considered the gold standard when the specificity of the STA method was evaluated.
STA was prepared as follows. Firstly, a basal medium was prepared dissolving 10g of peptone, 20g of glucose and 20g of agar (all from Britania Laboratory, Argentina) in 990ml of distilled water. The basal medium was sterilized at 121°C for 15min. Meanwhile, a stock solution of 2,3,5-triphenyltetrazolium chloride (TTZ) (Sigma–Aldrich, Argentina) was prepared dissolving 1g in 100ml of distilled water. This last solution was filter-sterilized. Finally, the basal medium was cooled at 55°C in a water bath and 10ml of the TTZ stock solution were added, reaching a 0.1g/l TTZ final concentration. Then, the media was plated in 90mm Petri dishes.
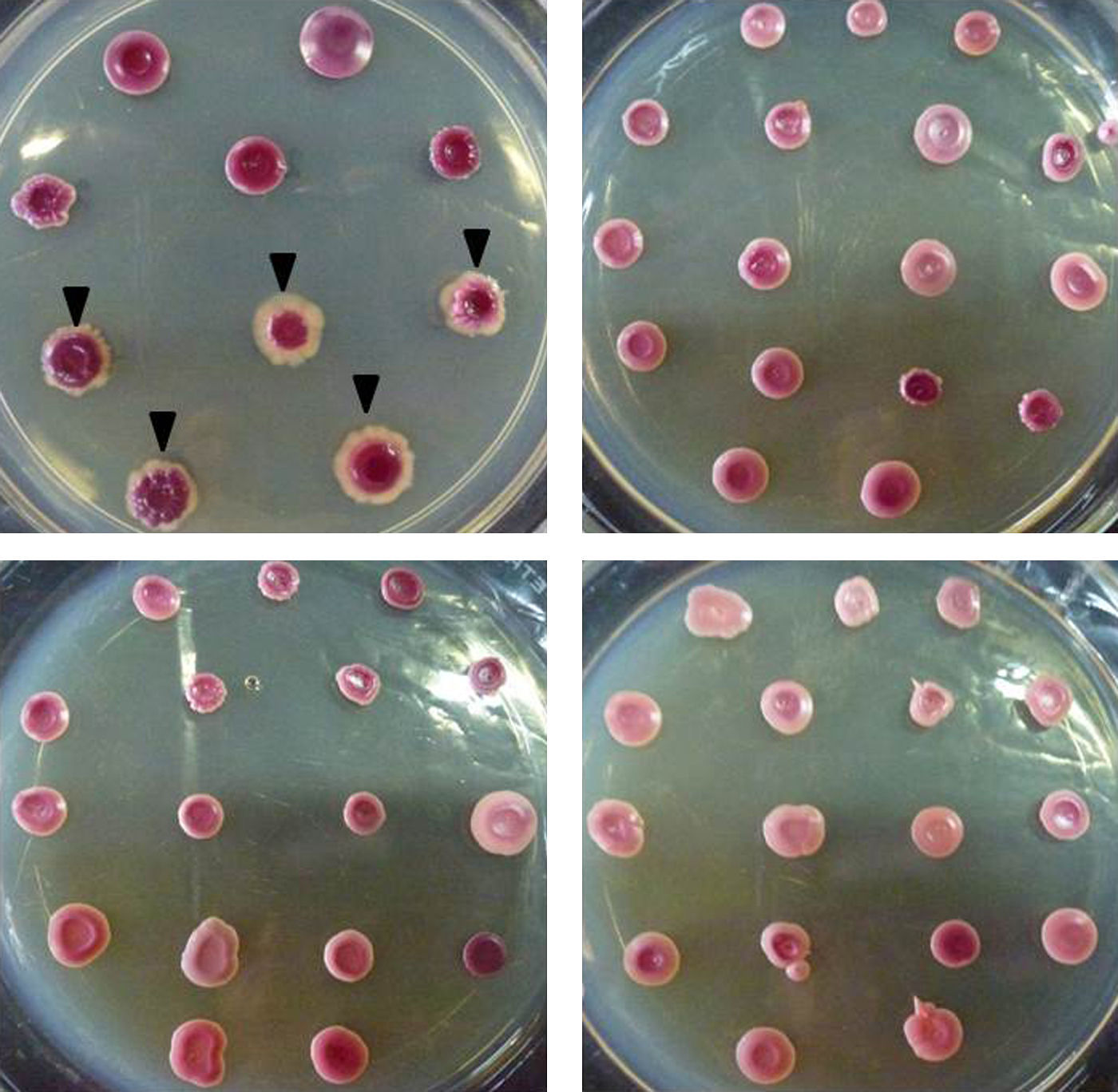
For the morphotype evaluation, fresh 24h-cultures in Sabouraud dextrose agar (peptone 1%, glucose 2%, agar 2%) were used. Four to five colonies were picked to obtain a 0.5 McFarland inoculum in water. Afterwards, 3μl of each cell suspension were inoculated onto STA plates (16 strains per plate), and incubated for 7 days at 28°C. The morphotype data collected included color (pink or violet), presence or absence of mycelial halo and colony texture (smooth or rough). The morphotypes were named using the nomenclature published by Quindós et al.11 The experiments were performed in triplicates in three separate days.
The 113 C. albicans strains showed three different morphotypes: pink with no mycelial halo (n=65, 57.5%), pink with mycelial halo (n=39, 34.5%), and violet with no mycelial halo (n=9, 8%). On the other hand, all the C. dubliniensis (n=13) strains produced violet colonies with large white mycelial halo (Fig. 1). Colony texture was not informative since C. albicans and C. dubliniensis showed both phenotypes. The morphotypes were reproducible even after a −86°C storage and multiple subcultures (data not shown).
The reduction of TTZ has been used as an aid for Candida identification since the 1980s.5 In 1992, Quindós et al. reported the use of colony morphotype on STA as a Candida species identification tool.11 In that report, 93.67% and 3.6% of the strains identified phenotypically as C. albicans were pink or violet, respectively, and only 2% of the C. albicans isolates showed violet colonies with mycelial halo. It would be possible that those results represented a 2% incidence of C. dubliniensis in the yeast collection used. This incidence was not reported since C. dubliniensis was proposed as separate species three years later.
In 1998, the reduction of TTZ was suggested as a useful tool for the differentiation of C. albicans from C. dubliniensis by Velegraky and Logotheti.19 However, in the aforementioned work, another basal medium was used; there is no description of the number of isolates tested and how the authors identified the strains as C. albicans or C. dubliniensis. Accordingly, the specificity and sensitivity evaluation was not performed. Also, Velegraky and Logotheti evaluated the color of the colony as the only characteristic. In the present study, we evaluated three morphotype characteristics on STA as a tool for C. dubliniensis – C. albicans differentiation using a total of 126 strains (122 isolated from different clinical sources) identified by two different molecular-based techniques. The method described here has 100% specificity and sensitivity when color and mycelial halo is considered together (all the C. dubliniensis showed violet colonies with large white mycelial halo). It has to be highlighted that we do not propose this test as a primary isolation media capable to distinguish C. dubliniensis and C. albicans directly from biological samples since other Candida species (e.g. C. tropicalis) are also able to reduce TTZ salts.18 We suggest the use of STA medium as and inexpensive, easy to perform differentiation tool for C. albicans and C. dubliniensis after a germ tube evaluation or starting from a green colony in Chromagar®Candida. Also, the seven days of incubation needed to see the result is an important disadvantage as a useful method in a clinical laboratory. However, STA medium would be useful for large epidemiology studies in reference labs since 16 strains could be studied per each 90mm Petri dish.
This work was financially supported in part by grants CAI+D prog. RH and PEIS 2011 both from the Universidad Nacional del Litoral (Argentina) to G.G.E. and S.G., respectively. C. Dudiuk has a predoctoral fellowship from Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET).