Sporotrichosis is a fungal infection caused by the Sporothrix schenckii complex. In order to colonize the host, the pathogen must neutralize the reactive oxygen species produced by the phagocytic cells during the respiratory burst. Little is known about these mechanisms in S. schenckii.
AimsTo identify the proteins differentially expressed after the exposure of S. schenckiisensu stricto to different concentrations of H2O2.
MethodsYeast cells of S. schenckiisensu stricto were exposed to increasing concentrations of H2O2. Proteins differentially expressed in response to oxidative stress were analyzed using two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) and identified by MALDI-MS/MS. RT-PCR assays were performed to evaluate the transcription of genes of the identified proteins.
ResultsConcentrations of H2O2 as high as 800mM allowed cell growth, and 200mM and 400mM were selected for comparative analysis by 2D-PAGE. This analysis revealed at least five differentially expressed proteins, which were identified as heat shock 70kDa protein (Hsp70), chaperonin GroEL, elongation factor 1-β (EF1-β), a hypothetical protein, and mitochondrial peroxiredoxin (Prx1). RT-PCR revealed that the transcription of the genes coding for some of these proteins are differentially regulated.
ConclusionsBased on these results, it is proposed that these proteins may be involved in the resistance of S. schenckii to oxidative stress, and play an important role in the fungus survival in the host.
La esporotricosis es una infección fúngica causada por el complejo Sporothrix schenckii. Para colonizar al huésped, los patógenos deben neutralizar las especies reactivas de oxígeno producidas por las células fagocíticas durante el estallido respiratorio. Poco se conoce sobre este mecanismo en S. schenckii.
ObjetivosIdentificar proteínas diferencialmente expresadas durante la exposición de S. schenckiisensu stricto a diferentes concentraciones de H2O2.
MétodosLevaduras de S. schenckiisensu stricto fueron expuestas a concentraciones crecientes de H2O2. Las proteínas diferencialmente expresadas en respuesta el estrés oxidativo fueron analizadas mediante electroforesis en geles de poliacrilamida en doble dimensión (2D-PAGE) e identificadas por MALDI-MS/MS. Se realizaron ensayos de RT-PCR para evaluar la transcripción de genes de las proteínas identificadas.
ResultadosConcentraciones altas de H2O2 (800mM) permitieron el crecimiento celular, y se seleccionaron las concentraciones de 200 y 400mM para el análisis comparativo mediante 2D-PAGE. Este análisis reveló al menos cinco proteínas diferencialmente expresadas, identificadas como proteína de choque térmico de 70 kDa (Hsp70), chaperonina GroEL, factor de alargamiento 1-β (EF1-β), una proteína hipotética y peroxirredoxina mitocondrial (Prx1). La RT-PCR reveló que la transcripción de los genes que codifican para algunas de estas proteínas se regula diferencialmente.
ConclusionesCon estos resultados pensamos que estas proteínas podrían estar involucradas en la resistencia de S. schenckiisensu stricto al estrés oxidativo y jugar un papel importante en la supervivencia del hongo en el huésped.
Sporotrichosis is a subcutaneous mycosis caused by the Sporothrix schenckii complex, a thermodimorphic fungus which is endemic in tropical and subtropical areas of Latin America. This disease has been reported as endemic because of the widespread use of immunosuppressive therapy, cancer, alcoholism and the incidence of acquired immunodeficiency syndrome (AIDS).17 Phenotypic and molecular studies have shown that S. schenckii is a complex of different cryptic species, some of which are considered of medical importance, such as S. schenckii sensu stricto, Sporothrix brasiliensis, Sporothrix globosa, Sporothrix mexicana, Sporothrix luriei and Sporothrix pallida.2,17–19
As other pathogens, the first barrier that Sporothrix must overcome to colonize its host is the immune system itself. Upon interaction with pathogens, phagocytes produce reactive oxygen species (ROS) during the oxidative burst, which are essential components of the immune response against the invading microorganisms.4,27 ROS are mainly formed in the mitochondrial electron transport chain by the partial reduction of O2, generating superoxide anions (O2•−), hydrogen peroxide (H2O2) and hydroxyl radicals (HO•). Pathogens have developed mechanisms involving enzymatic and non-enzymatic systems that enable them to detoxify ROS and evade phagocytic cells.27 After the phagocytosis of S. schenckii, monocytes and macrophages are strongly induced to produce ROS.25 It has been shown that in this organism, the superoxide radical has fungistatic and fungicidal activity, and its absence is associated with higher mortality in experimental mouse infections.14 Despite the relevance of this anti-oxidant activity, there are few studies dealing with the responses of Sporothrix to oxidative stress (OS).22 It is well documented that the response to OS (OSR) depends on the phase growth of the pathogen as it has been demonstrated that exponentially growing cells are more susceptible than those in the stationary phase of growth.6,8,21,31 A recent work in this laboratory has focused on the mechanisms used by S. schenckii sensu stricto to respond to OS in the stationary phase of growth. This phase is of special interest as cells remain in quiescent state and very much emulate their lifestyle in nature.21,32 Quiescent cells represent almost 60% of earth biomass and they can survive for long periods of time, sometimes years, in the absence of nutrients, a feat of astonishing resilience.11 It has been reported that in pathogenic organisms such as some Candida species, the stationary phase favours colonization of the human host where they remain quiescent until optimal nutrient conditions exist to infect and invade it. S. schenckii exhibits a similar behaviour. While in some Candida species some proteins involved in the OSR have been identified,7,23 the same is not true for S. schenckii complex.
On this background, we considered it relevant to identify proteins potentially involved in the OSR following exposure of stationary phase yeast cells of S. schenckii sensu strictu to H2O2. We identified at least five proteins that were up- or down-regulated after exposing the pathogen to increasing concentrations of H2O2. RT-PCR revealed that transcription of genes coding for some of these proteins are differentially regulated. We propose that these proteins are involved in the mechanisms of resistance against ROS and may contribute to the survival of the fungus in the host.
Material and methodsStrain and culture conditionsThroughout this study, the S. schenckii ATCC 58251 strain was used. To obtain the yeast morphotype cells in the stationary-phase, 500-ml Erlenmeyer flasks containing 200ml of brain heart infusion medium (BHI, Difco) were inoculated with 5×105conidia·ml−1 and incubated at 37°C for 8 days on a rotary shaker at 120rpm. Yeast cells were harvested by centrifugation at 7000×g for 10min and used for the assays described below.
Assays of H2O2 susceptibilityS. schenckii was grown for 8 days at 37°C. To assay H2O2 susceptibility, yeast cells were diluted in fresh BHI medium to an OD600nm of 0.5 in sterile deionized water. The cultures were divided in equal parts, exposed to different H2O2 concentrations (0 to 1000mM) and shaken (120rpm) at 37°C. After 60min, an aliquot was taken from the cultures treated with the oxidant, adjusted to an OD600nm of 0.5 and used to prepare exponential dilutions in 96-well boxes. Each dilution was spotted onto YPG plates (0.3% yeast extract, 1% peptone, and 2% glucose) and incubated at 37°C for 48h. The experiments were carried out thrice. These protocols were adapted from previous studies with other organisms.6,7
Extraction of proteinsNon-exposed and H2O2-exposed cell cultures were centrifuged at 13,000×g at 4°C for 15min and the supernatant was carefully discarded. The cell pellet was washed thrice with washing buffer [200mM Tris–HCl, pH 8.5, 1mM phenylmethylsulfonyl fluoride (PMSF)] by centrifugation at 7000×g at 4°C for 10min. Washed cells were resuspended in lysis buffer (200mM Tris–HCl, pH 8.5, 1mM PMSF, 200mM NaCl, 0.5% SDS, 25mM EDTA) and broken with glass beads (0.45–0.5mm in diameter) by alternate periods of breaking (40s) and cooling (60s) until all cells were broken. The cell homogenate was centrifuged at 7000×g at 4°C for 10min. The supernatant was carefully aspirated with a Pasteur pipette and the cell pellet was discarded. Proteins in the supernatant were precipitated with 70% (v/v) ethanol at −20°C for 3h and stored at −70°C until further use. Protein concentration was determined by the DC method (Bio-Rad) using bovine serum albumin (BSA) as standard.
2D-PAGE and image analysis2D-PAGE was performed using 7-cm strips with an immobilized 4–7 pH gradient (Bio-Rad) as described by Ruiz-Baca et al.26 Briefly, samples containing 80μg of total protein were cleaned using the cleaning kit from Bio-Rad (2D Clean-Up Kit) as described above and resuspended in 140μl of rehydration buffer [7M urea, 2M thiourea, 4% CHAPS, 50mM dithiothreitol (DTT), 0.2% ampholytes (Biolyte 3/10) and 0.001% bromophenol blue]. After rehydration for 16h, the strips were subjected to isoelectric focusing with voltage gradients of 0 to 250V for 15min, 250 to 4000V for 2h, and 4000V to completion at 8500V/h. After isoelectric focusing, the strips were incubated sequentially for 15min in equilibrium buffer I (0.375mM Tris/HCl, pH 8.8, 6M urea, 20% glycerol, 2% SDS and 0.5% DTT) and equilibrium buffer II (0.375mM Tris/HCl, pH 8.8, 6M urea, 20% glycerol, 2% SDS and 2% iodoacetamide) under constant stirring. For the second dimension, the strips were placed on top of a 10% SDS-PAGE gel and covered with a 0.5% agarose overlay. The proteins were separated at 95V for 2h in a Mini-Protean 3 system (Bio-Rad) and then stained with colloidal Coomassie blue. Images were captured with a ChemiDocTM XRS+System (Bio-Rad) and analyzed with a PDQuest™ 2-D (Bio-Rad) software. Comparisons of the gels were done using a synthetic image containing all the protein spots of analyzed gels. The intensity of the spots was normalized and validated in the master gel. A spot was considered relevant when there was a minimum of a two-fold difference in its intensity as compared with the corresponding spot obtained from the oxidant-untreated sample.
Protein identificationSpots of interest were manually excised from 2D electrophoresis gels. The gel pieces were destained and enzymatically digested according to the modified protocol of Shevchenko et al.30 Resulting tryptic peptides were concentrated to an approximate volume of 10μl. Nine μl of this sample were loaded onto a ChromXP Trap Column C18-CL precolumn (Eksigent, Redwood City CA); 350μm×0.5mm, 120A° pore size, 3μm particle size and desalted with 0.1% trifluoroacetic acid (TFA) in H2O at a flow rate of 5μl/min during 10min. Then, peptides were loaded and separated on a Waters BEH130 C18 column (Waters, Milford, MA); 100μm×100mm, 130A° pore size, 1.7μm particle size, using a HPLC Ekspert nanoLC 425 (Eksigent, Redwood City CA) with 0.1% TFA in H2O and 0.1% TFA in acetonitrile (ACN) as mobile phases A and B, respectively, under the following lineal gradient: 0–3min 10% B (90% A), 60min 60% B (40% A), 61–64min 90% B (10% A), 65 to 90min 10% B (90% A) at a flow rate of 250nl/min. Eluted fractions were automatically mixed with a solution of 2mg/ml of α-cyano-4-hydroxycinnamic acid (CHCA) in 0.1%TFA and 50% ACN as a matrix, spotted in a stainless steel plate of 384 spots using a MALDI Ekspot (Eksigent, Redwood City CA) with a spotting velocity of 20s per spot at a matrix flow rate of 1.6μl/min. The spots generated were analyzed by a MALDI-TOF/TOF 4800 Plus mass spectrometer (ABSciex, Framingham MA). Each MS Spectrum was acquired by accumulating 1000 shots in a mass range of 850–4000Da with a laser intensity of 3700. The 100 more intense ions with a minimum signal-noise of 20 were programmed to fragmenting. The MS/MS spectra were obtained by fragmentation of selected precursor ions using collision induced dissociation and acquired by 3000 shots with a laser intensity of 4400. Generated MS/MS spectra were compared using a Protein Pilot software v. 2.0.1 (ABSciex, Framingham, MA) against S. schenckii strain ATCC 58251 and 1099-18 database (downloaded from Uniprot; 8673 and 10292 protein sequences, respectively) using Paragon algorithm. The search parameters were carbamidomethylated cysteine, trypsin as a cut enzyme, all the biological modifications and amino acid substitution set by the algorithm, as well as phosphorylation emphasis and Gel-based ID as special factors. The detection threshold was considered in 1.3 to acquire 95% of confidence. The identified proteins were grouped by ProGroup algorithm in the software to minimize redundancy.
Reverse transcription-PCR (RT-PCR)Total RNA from Sporothrix cells was isolated using the Trizol reagent (Invitrogen) according to the manufacturer's instructions, and DNase I (Invitrogen) was used to eliminate DNA contamination. Synthesis of cDNA and PCR were carried out as described elsewhere using the ImProm-IITM Reverse Transcription System (Promega). The expression levels of prx1 and hsp70 genes were normalized by β-tubulin gene (tub). The RT primers used for each gene are shown in Table 1. Synthesis of cDNA was carried out at 42°C for prx1, hsp70 and tub genes and PCR was performed at 58°C for the three analyzed genes. PCR amplification of DNase-treated RNA was performed without the reverse-transcription procedure (No-RT). Lack of amplification of PCR products confirmed the complete elimination of DNA from RNA samples. The PCR products were visualized on agarose gel stained with ethidium bromide. The quantification of PCR products was carried out using an ImageJ1.51j8 software (Wayne Rasband, National Institutes of Health, USA).
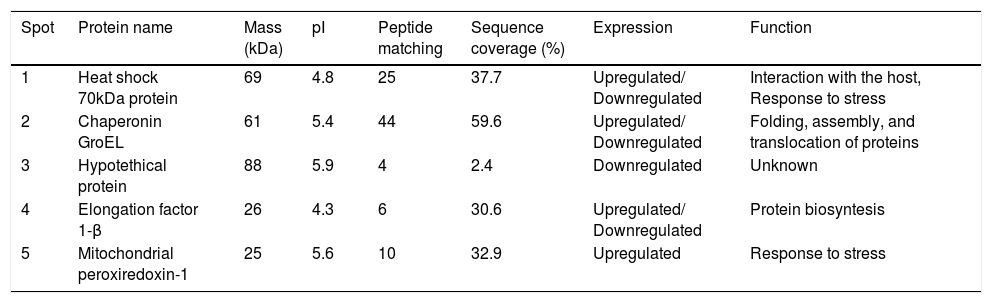
Proteins separated on 2D-PAGE gels and analyzed by MALDI-MS/MS.
| Spot | Protein name | Mass (kDa) | pI | Peptide matching | Sequence coverage (%) | Expression | Function |
|---|---|---|---|---|---|---|---|
| 1 | Heat shock 70kDa protein | 69 | 4.8 | 25 | 37.7 | Upregulated/ Downregulated | Interaction with the host, Response to stress |
| 2 | Chaperonin GroEL | 61 | 5.4 | 44 | 59.6 | Upregulated/ Downregulated | Folding, assembly, and translocation of proteins |
| 3 | Hypotethical protein | 88 | 5.9 | 4 | 2.4 | Downregulated | Unknown |
| 4 | Elongation factor 1-β | 26 | 4.3 | 6 | 30.6 | Upregulated/ Downregulated | Protein biosyntesis |
| 5 | Mitochondrial peroxiredoxin-1 | 25 | 5.6 | 10 | 32.9 | Upregulated | Response to stress |
Mass spectra were analyzed with Protein Pilot software v. 2.0.1 (ABSciex, Framingham MA) against S. schenckii. Protein name: protein name as deduced by comparing peptide sequences via the software BLAST. Molecular weight (kDa): theoretical molecular mass predicted from the amino acid sequence of the identified protein. pI: theoretical isoelectric point predicted from the amino acid sequence of the identified protein. Sequence coverage: coverage of the amino acid sequence of the identified protein. Peptide matching: number of matched peptides based on MS/MS data searching, excluding the duplicate matches.
The quantification of PCR products were analyzed by a single-factor ANOVA in a completely randomized design with three levels of peroxide concentration. Then, a Tukey post hoc multiple comparison of means with a 95% family-wise confidence level was performed. A p<0.05 was considered statistically significant. These analyses were performed using R, a programming language for statistical computing.24
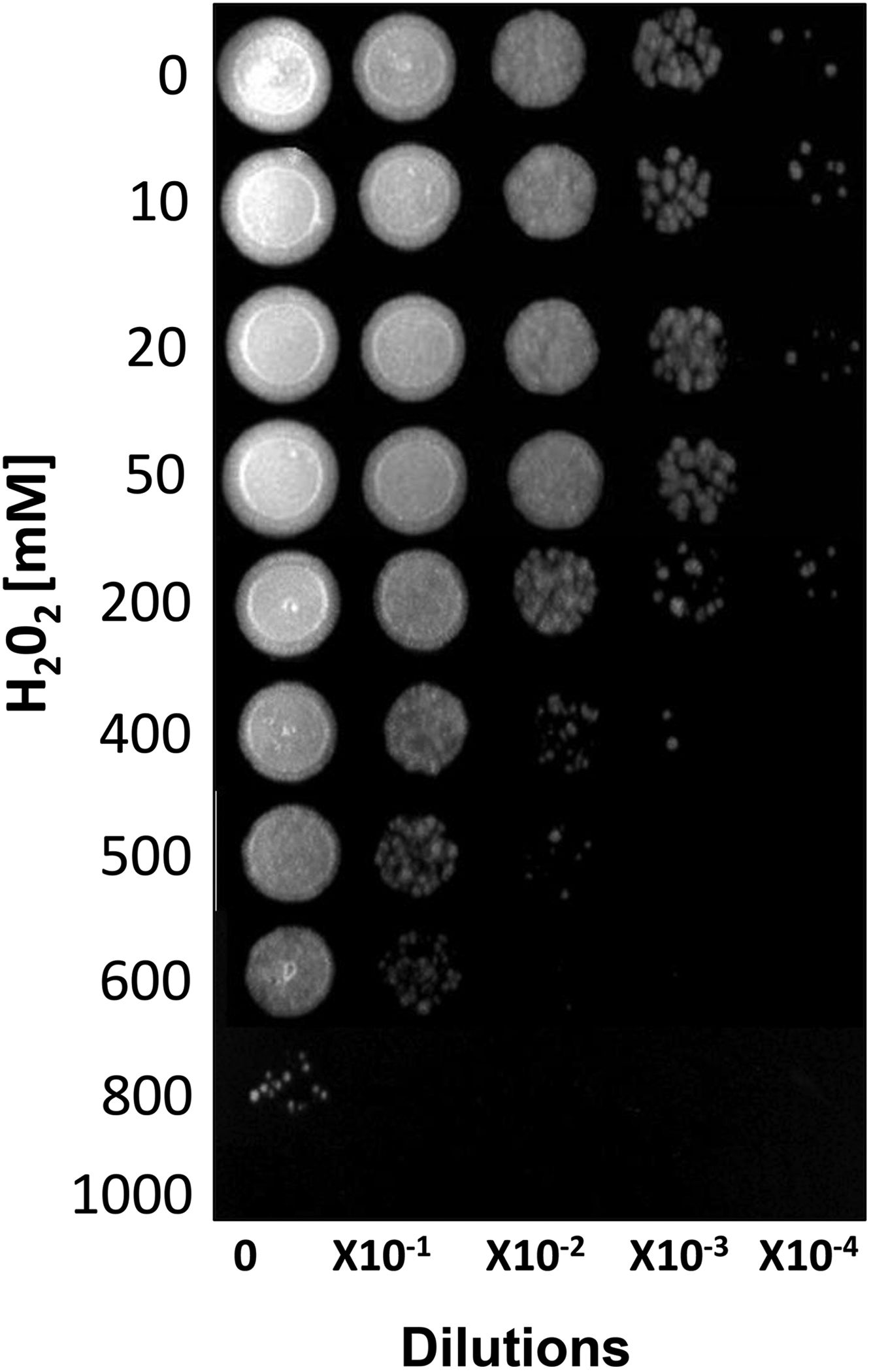
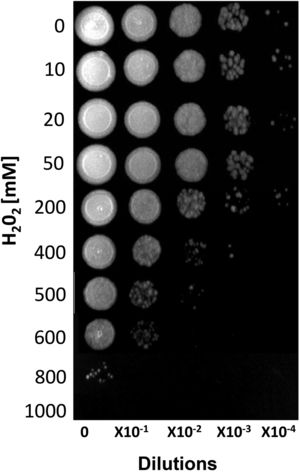
ResultsSusceptibility to H2O2To increase our knowledge of the resistance mechanisms of S. schenckiisensu stricto, we considered it relevant to identify some proteins presumptively involved in OSR. To this purpose, stationary-phase cultures of yeast cells of the fungus were exposed to increasing concentrations of H2O2 (Fig. 1). Fungal growth was not affected when cell dilutions were exposed to concentrations between 0 and 50mM of H2O2. At 200mM, inhibition was observed at a cell dilution of 1×10−2. At 400mM and at a dilution of 1×10−1, fungal growth decreased and was fully inhibited at all higher cell dilutions with no significant differences at 500mM. At 600mM, growth decreased at dilution 0, with poor growth at 800mM and full inhibition at 1000mM.
Susceptibility of S. schenckii sensu stricto to H2O2. Cultures of stationary-phase yeast cells (OD600nm 0.5) were incubated under constant stirring in the presence of the indicated concentrations of H2O2 at 37°C. Samples of these suspensions were exponentially diluted in 96-well plates and each dilution was spotted onto YPG plates that were incubated at 37°C. Growth was inspected after 48h.
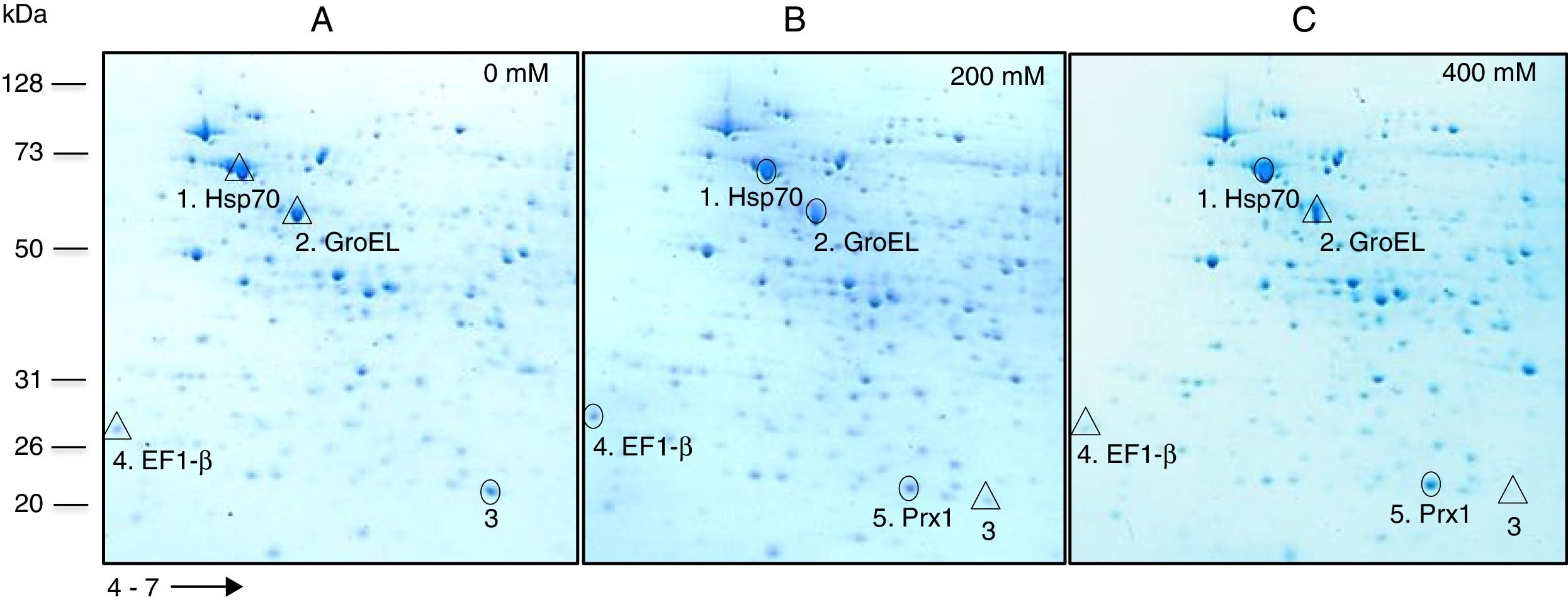
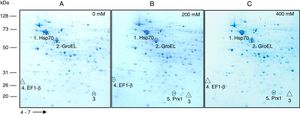
Based on their effect on growth, concentrations of 0, 200 and 400mM H2O2 were selected to carry out a comparative analysis of the total protein expression by S. schenckii sensu stricto. Analysis by 2D-PAGE at different concentrations of H2O2 showed the presence of at least five differentially expressed proteins (Fig. 2) which were identified as a heat shock 70kDa protein (Hsp70), chaperonin GroEL, elongation factor 1-β (EF1-β), a hypothetical protein and mitochondrial peroxiredoxin (Prx1) (Table 1). Accordingly, an upregulated expression of Hsp70 and Prx1 was observed (Fig. 2; spots 1 and 5, respectively). Other proteins that were either up- or downregulated were chaperonin GroEL and EF-1β (Fig. 2; spots 2 and 4, respectively). Also, a downregulated expression of a hypothetical protein was observed in response to OS (Fig. 2; spot 3).
Analysis of total protein extracts from yeast cells of S. schenckiisensu stricto by 2D-PAGE in a pH 4–7 gradient in gels stained with colloidal Coomassie Blue, after exposure to 0 (A), 200 (B) and 400 (C) mM H2O2. Spots marked with circles and triangles correspond to up- and down-regulated proteins, respectively, with respect to the reference condition. kDa, kilodalton.
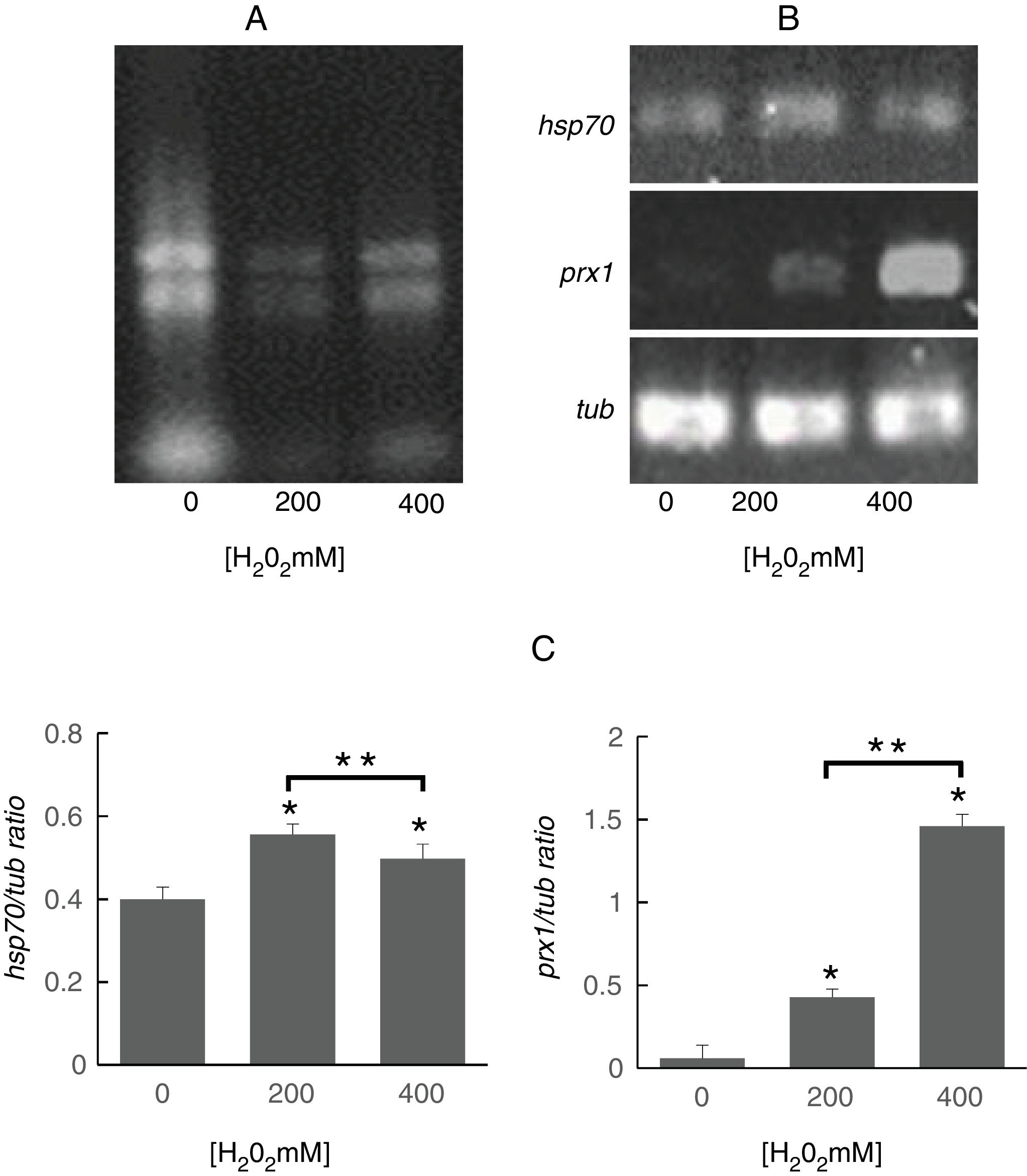
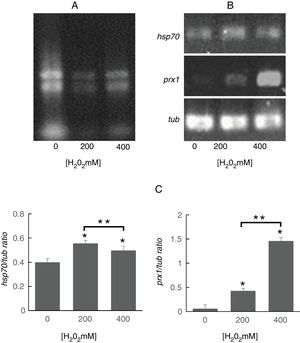
To determine whether the regulation of identified proteins occurred at the transcriptional level, we evaluated the expression of genes encoding Prx1 and Hsp70. To this purpose, it was important to assess the quality of the RNA obtained from S. schenckii sensu stricto as it has been reported that ROS can damage nucleic acids.4 It was observed that the ribosomal RNA bands were well defined indicating that it was suitable for analysis (Fig. 3A). Following exposure to H2O2, significant differences were found in the expression of the prx1 and hsp70 genes at 200 and 400mM H2O2 (p<0.05) as compared with the control. On the other hand, prx1 showed a significant increase at 400mM H2O2 compared with 200mM H2O2 (p<0.05), while hsp70 showed a significant decrease at 400Mm H2O2 compared with 200mM (p<0.05) (Fig. 3B and C).
Analysis of prx1 and hsp70 gene expression by RT-PCR. (A) Total RNA of S. schenckii sensu stricto exposed to the indicated concentrations of H2O2. (B) RT-PCR products from the analysis of hsp70, prx1 and tub gene expression in S. schenckii sensu stricto. (C) Densitometry of the bands was measured using the ImageJ 1.51j8 software. The quantification of prx1 and hsp70 was perfomed. Data are presented as the mean±SD, n=3. *Significant differences as compared to control (p<0.05). **Significant differences between concentrations of H2O2 (p<0.05).
Identification of proteins potentially involved in the resistance of S. schenckiisensu stricto to oxidant agents is a useful approach to understand how the fungus can protect itself from OS. Here, stationary-phase yeast cells of the fungus were subjected to OS with different concentrations of H2O2. This oxidant started to affect the growth of S. schenckii at 200–400mM, the inhibition increased at 600mM and no cells remain viable at 1000mM. In other pathogenic fungi such as Paracoccidioides brasiliensis and Candida species, the response to oxidative stress in the growth phase was different.1,8,23 Yeast cells of P. brasiliensies in the stationary phase were more resistant to H2O2 than their counterparts in the exponential phase after exposure to 0–100mM H2O2. The survival rates of cells from both phases exceeded 70% with all tested concentrations of H2O2.8 In contrast, species of the Candida genus in the stationary phase of growth reveals that C. albicans is more susceptible to H2O2 as its growth was completely inhibited at 300mM. Other species such as C. glabrata and C. parapsilosis survived at 500mM, while C. krusei did so at 1500mM.23 Resistance of Candida species to H2O2 has also been observed by other authors,1 thus reinforcing the notion that pathogenic organisms have developed mechanisms to detoxify H2O2 that enable them to evade the immune system and colonize the host.
2D-PAGE of extracts from S. schenckii sensu stricto yeast cells exposed to two concentrations of H2O2 showed the presence of at least five differentially expressed proteins. These were identified as Hsp70, chaperonin GroEL, a hypothetical protein, EF-1β and mitochondrial Prx1 (Fig. 2, spots 1–5, respectively. See also Table 1). Fungal Hsp70 has been implicated in various cellular functions, including OS, cell adhesion, macrophage activation, receptor expression and macromolecule internalization.15,28 Studies in species of Candida,7P.brasiliensies9 and Cryptococcusneoformans25 suggest an important role of Hsp70 in the interaction with host cells and in the response to OS. On the other hand, the chaperonin GroEL in bacteria is required for the proper folding of many proteins. Studies indicate that these chaperones are overexpressed under conditions of cellular stress, including heat, so they are considered as heat shock proteins5. On its part, EF-1β plays an important role in protein synthesis. Accordingly, McGoldrick et al.20 reported 67 sequences of glutathione transferase-like proteins encoded in 21 fungal species, some related to EF1-β, suggesting their involvement in the ROS. Also, the differentiated expression of EF1-β was noted in Candida species exposed to H2O2 and menadione, reinforcing its role in the stress response, perhaps as a modulator of transcription of other proteins.23
Another protein identified here was a mitochondrial Prx1. Fungal peroxiredoxins are ubiquitous enzymes that protect cells against OS.12,29 An in silico analysis by Ortega et al.22 demonstrated that both S. brasiliensis and S. schenckiisensu stricto have four putative peroxiredoxins in their genome, but to date it is unknown which of these genes are modulated by H2O2. In accordance with our results, a proteomic analysis in Aspergillus fumigatus revealed the differential expression of 28 proteins, including Prx1, which increased their concentration after 45min of exposure to H2O2.16 In the same line, Kusch et al.15 exposed C. albicans to two oxidizing agents, H2O2 and diamide, and observed the differential expression of 52 proteins, including Ipf2431p, which is homologous to Tsa1p in S. cerevisiae where it plays the role of a peroxiredoxin. Another study in Candida species exposed to H2O2 and menadione revealed the differential expression of 15 moonlight-like cell wall proteins including Tsa1p,23 while in S. cerevisiae it was proposed that Prx1 acts as a redox signalling molecule that oxidizes Trx3. At high concentration oxidized Trx3 can produce apoptosis, indicating that when Prx1 is unable to detoxify reactive mitochondrial oxygen species it induces apoptosis to remove the affected cells.13 Further studies by proteomics and bioinformatic approaches about the response to OS in exponentially and stationary-phase in Sporothrix will be necessary to get a deeper knowledge of the defense mechanisms of this pathogen as they occur in the phagocyte.
To determine whether the regulation of the identified proteins occurred at the transcriptional level, the expression of the genes encoding Hsp70 and Prx1 was evaluated. To this purpose, it was important to assess the quality of the RNA obtained from S. schenckiisensu stricto as it has been reported that ROS can damage nucleic acids4. The ribosomal RNA bands were well defined indicating that it was suitable for analysis (Fig. 3A). Following the exposure of S. schenckii sensu stricto to H2O2, significant differences were found (p<0.05) in the expression levels of prx1 and hsp70 genes. Thus, while a significant increase in the expression of prx1and hsp70 were observed at 200 and 400mM H2O2 compared with the control, a significant decrease in hsp70 occurred at 400mM H2O2 compared with 200mM H2O2 (Fig. 3B and C). These results are in agreement with those obtained from the proteomic analysis carried out after the exposure of S. schenckii sensu stricto yeast cells to ROS (Fig. 2), therefore indicating that changes observed for the proteins identified in this study are regulated at the transcriptional level.
Comparative studies of the virulence of S. schenckii complex demonstrated that S. brasiliensis was the most virulent followed by S. schenckii sensu stricto, S. globosa, S. mexicana y S. pallida.3 Fernandes et al.10 evaluated the secretion profiles of proteins from different isolates of S. schenckii sensu stricto, S. brasiliensis and S. globosa, and reported the expression of different proteins. They also observed that the humoral response of animals infected with these species was different. The most virulent isolates shared two common antigens of 60kDa and 110kDa, which are likely to be involved in virulence. In the same line, Ortega et al.22 observed that S. brasiliensis was more resistant to stress by peroxides as compared with S. schenckii sensu stricto, suggesting that the S. schenckii species exhibit different strategies for adaptation, depending on the route of infection, a phenomenon most likely related with virulence.
ConclusionsBased on the results presented in this study, we propose that the identified proteins are likely to be involved in the resistance of this fungus against OS, helping the pathogen to evade the host phagocytic cells, reach the blood stream and cause an invasive mycosis.
Conflict of interestsThe authors declare that there were no conflicts of interest with any organization or entity with a financial interest, or financial conflict with the material discussed in this work.
The authors appreciate the technical assistance and facilities provided for the analysis of MALDI-MS/MS by M.C. Emmanuel Ríos Castro of the Unidad de Genómica, Proteómica y Metabolómica (LaNSE; CINVESTAV-Unidad Ciudad de México, México). We are grateful to M.C. Omar Posada-Villarreal for his support in the statistical analysis. This work was supported by grant CB-2011 No.167737 from the Consejo Nacional de Ciencia y Tecnología (CONACyT, México) to ERB.