Cryptococcosis is a fungal infection with a worldwide distribution, mainly caused by Cryptococcus neoformans and Cryptococcus gattii.
AimsTo molecularly characterize the mating-types, serotypes, genotypes and antifungal susceptibility profiles of a set of retrospectively isolated C. neoformans strains from Lima, Peru.
MethodsA set of 32 Cryptococcus spp. strains from the Institute of Tropical Medicine of the National University of San Marcos, Lima, Peru, were included in this retrospective study. Twenty-four strains were isolated from patients, while the remaining 8 were isolated from the environment.
ResultsUsing conventional PCR, 27 (84.4%) of the isolates were identified as C. neoformans var. grubii mating-type alpha and serotype A. Using the AFLP fingerprinting, it was shown that 16 (50%) of the C. neoformans strains were genotype AFLP1, 13 (40.6%) were genotype AFLP1B, 2 (6.3%) were genotype AFLP2, and 1 (3.1%) was found to be a hybrid between both C. neoformans varieties (genotype AFLP3). The antifungal susceptibility profiles for amphotericin B, fluconazole and voriconazole showed that all the 32 C. neoformans are sensitive to these antifungal compounds.
ConclusionsIn this study we observed that C. neoformans var. grubii (AFLP1 and AFLP1B) and C. neoformans var. neoformans (AFLP2) were the only cryptococcal varieties involved. All strains were found to be sensitive to the antifungals tested, results that are consistent with those found in the international literature.
La criptococosis es una infección fúngica de distribución mundial, causada principalmente por Cryptococcus neoformans y Cryptococcus gattii.
ObjetivosDeterminar el tipo de apareamiento, los serotipos, los genotipos y la sensibilidad antifúngica de las cepas de C. neoformans provenientes de Lima Perú.
MétodosSe analizaron 32 cepas de Cryptococcus spp. del Instituto de Medicina Tropical de la Universidad Nacional Mayor de San Marcos de Lima, Perú. Veinticuatro cepas provenían de pacientes y 8 cepas fueron obtenidas de muestras ambientales.
ResultadosMediante la técnica de reacción en cadena de la polimerasa se determinó que 27 (84,4%) aislamientos eran C. neoformans var. grubii, serotipo A, con el tipo de apareamiento alfa. Asimismo, empleando la técnica AFLP, se determinó que 16 (50%) cepas de C. neoformans eran del genotipo AFLP1 y 13 (40,6%) del genotipo AFLP1B; además, 2 (6,3%) eran del genotipo AFLP2 (C. neoformans var. neoformans) y 1 (3,1%) resultó ser un híbrido entre ambas variedades (AFLP3). Los perfiles de sensibilidad antifúngica para anfotericina B, fluconazol y voriconazol mostraron que las 32 cepas de C. neoformans eran sensibles al panel de antifúngicos.
ConclusionesEn el presente estudio observamos que C. neoformans var. grubii (AFLP1 y AFLP1B) y C. neoformans var. neoformans (AFLP2) fueron las variedades encontradas. Además, todas las cepas resultaron sensibles al panel de antifúngicos, en concordancia con la literatura internacional.
The genus Cryptococcus includes more than 100 species, of which only the Cryptococcus neoformans/Cryptococcus gattii complex is considered a real human pathogen.23C. neoformans is divided into the two varieties C. neoformans var. grubii (serotype A; genotypes AFLP1, AFLP1A and AFLP1B), C. neoformans var. neoformans (serotype D; genotype AFLP2), and hybrids between both varieties (serotype AD; genotype AFLP3).3C. gattii was until a decade ago considered a variety of C. neoformans, but was finally raised to the species level based on physiological, ecological, epidemiological and genetic differences.24C. gattii can be divided into five genotypes: AFLP4 (serotype B), AFLP5 (serotype C), AFLP6 (serotype B), AFLP7 (serotype C) and AFLP10 (serotype B).19 Hybrids between C. gattii and C. neoformans have recently been described.1
C. neoformans var. grubii is globally distributed, and can mainly be found in soil contaminated with bird droppings.12C. neoformans var. neoformans is commonly encountered in Europe but less frequently observed to cause disease outside this continent. Both varieties primarily affect immunocompromised patients.30C. gattii is mainly found in tropical and subtropical climates, and has been isolated from a variety of natural sources, mostly affecting immunocompetent patients.16 The incidence of cryptococcosis in HIV-infected patients has decreased dramatically in developed countries after the HAART era; unfortunately, in developing countries without access to HAART the incidence is considerable.30 Moreover, in Peru, the majority of the patients with HIV are homosexual men; however, the incidence is increasing in heterosexuals. In 2000–2010, the prevalence of treated HIV-positive patients with cryptococcosis was 14% (259/1848) at the Dos de Mayo National Hospital (one of the largest in the country), being neurocryptococcosis the most frequent diagnosis.20 The incidence of neurocryptococcosis in HIV-infected patients in Peru before the HAART era was 5.6%10 and, as was observed globally, it decreased after the implementation of HAART. However, neurocryptococcosis still represents a public health problem. Despite the administration of HAART, some patients start the treatment in advanced stages of the disease, as in many cases the diagnosis of HIV-infection is performed when the patient presents an opportunistic disease such as cryptococcosis.
The objective of this study was to determine the mating-types, serotypes, genotypes and antifungal susceptibility profiles of C. neoformans strains available in the culture collection of the Institute of Tropical Medicine from the National University of San Marcos, Lima, Peru.
Materials and methodsA set of 32 Cryptococcus spp. strains from the Institute of Tropical Medicine from the National University of San Marcos (ITM-UNMSM), Lima, Peru, was included in this retrospective study. The ITM-UNMSM is one of the main centers for tropical infectious diseases research in Peru. The samples were collected between January 2008 and December 2012. Clinical samples came from the Dos de Mayo National Hospital in Lima, and the Daniel Alcides Carrion National Hospital in Callao. Twenty-four strains came from patients, while the remaining eight were years ago isolated from the local environment. Of the 24 clinical strains, 23 came from HIV-infected patients and one from an immunocompetent individual. In all the clinical cases, the samples were taken from the cerebrospinal fluid. The environmental strains came from pigeon droppings collected from around the university campus of the National University of San Marcos.
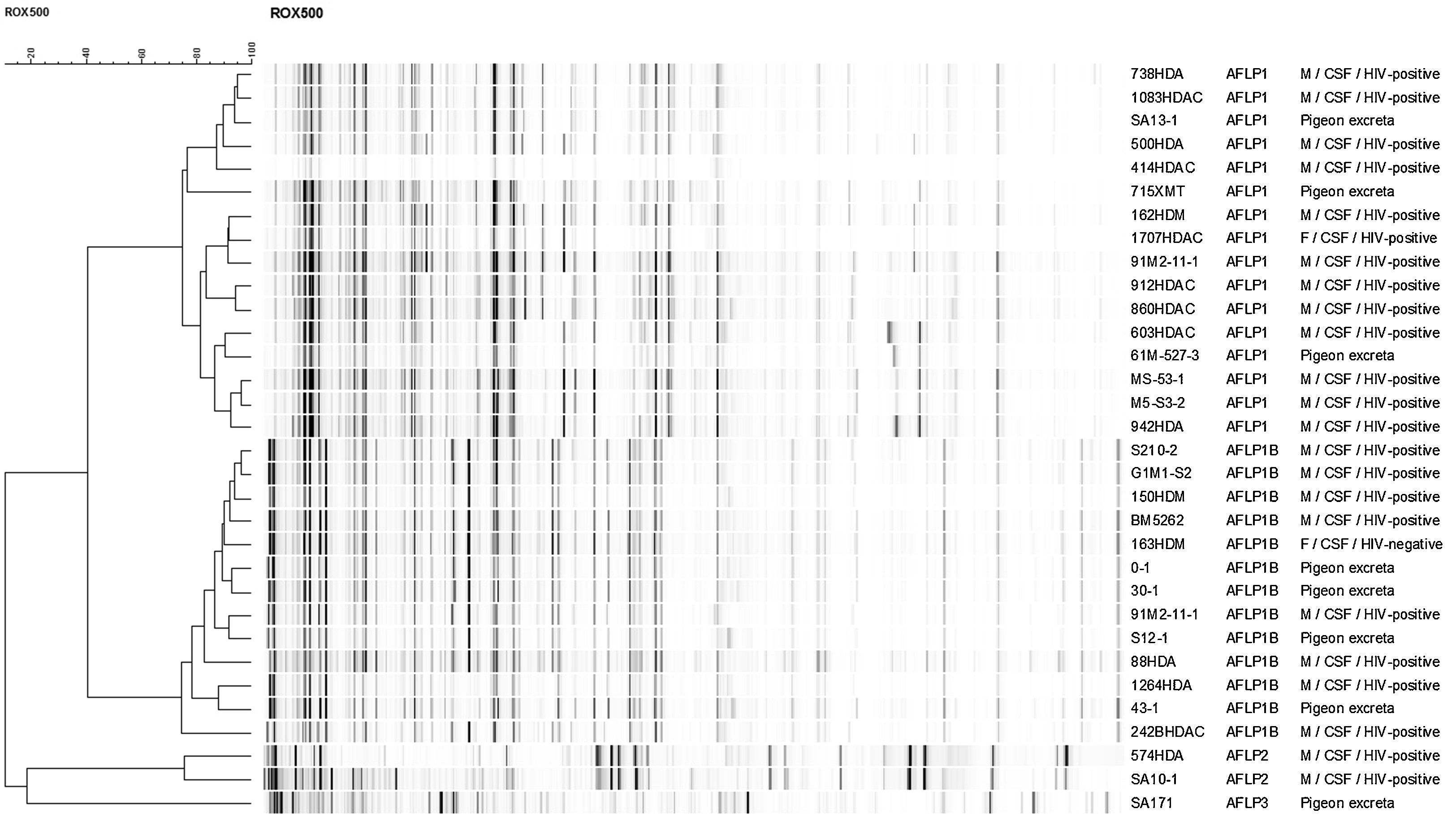
Mating-type, serotype and genotype determinationThe mating-type and serotype were molecularly determined using four conventional PCRs that specifically amplify the STE20a and STE20α allele of serotype A and D strains.2 The genotype was determined by using amplified fragment length polymorphism (AFLP) fingerprinting as previously described.17 The reference strains CBS8710 (AFLP1; αA), CBS9172 (AFLP1; aA), CBS11261 (AFLP1A; αA), CBS10083 (AFLP1B; αA), CBS10511 (AFLP2; αD), CBS10513 (AFLP2; aD), CBS10080 (AFLP3; αAaD), CBS10078 (AFLP4; αB), CBS10081 (AFLP5; αB), CBS10082 (AFLP6; αB), CBS10101 (AFLP7; αC) and IHEM14941 (AFLP10; aB) were included to determine the genotype. A dendrogram of the Peruvian strains was constructed using single linkage clustering in combination with the Pearson correlation using BioNumerics version 6.6 (Applied Maths, Sint-Martens-Latem, Belgium).
Antifungal susceptibilityThe antifungal susceptibility was evaluated by antifungal disk diffusion testing according to the Clinical and Laboratory Standards Institute.8 According to these guidelines, Mueller-Hinton agar supplemented with 2% glucose and 0.5μg/ml of methylene blue (MHM) was used. The antifungal panel consisted of (all from HiMedia Laboratories, Mumbai, India) fluconazole (25μg), voriconazole (1μg) and amphotericin B (20μg). The plates were incubated at 35°C. Quality controls were tested in the same way with Candida parapsilosis ATCC 22019 and Candida krusei ATCC 6528.
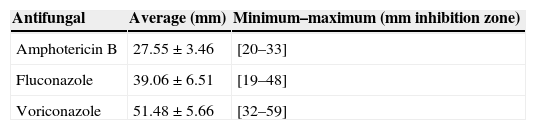
The inhibition zones were measured in millimeters at 48 and 72h. Breakpoints for fluconazole and voriconazole were determined according to Khan et al.22 The breakpoints for fluconazole were susceptible (S) ≥19mm, susceptible dose-dependent (SDD) 15–18mm, and resistant (R) ≤14mm. For voriconazole breakpoints were (S) ≥17mm, SDD 14–16mm, and R≤13mm. The set breakpoints for amphotericin B, according to Da Silva et al.,11 were S>10mm, R<10mm.
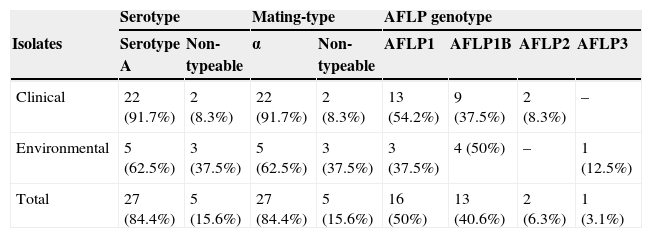
ResultsMating-types, serotypes and genotypesTwenty-seven (84.4%) strains out of the 32 strains tested were found to be alpha mating-type and serotype A by using the conventional PCR approach. This was confirmed by AFLP fingerprinting since these 27 strains were found to be C. neoformans var. grubii genotype AFLP1 (n=16; 50%) and AFLP1B (n=11; 34.4%). Five strains (15.6%) were untypeable by PCR but were found by AFLP fingerprinting to belong either to genotype AFLP1B (n=2; 6.2%), AFLP2 (n=2; 6.2%) or AFLP3 (n=1; 3.1%), representing C. neoformans var. grubii, C. neoformans var. neoformans, and the hybrid between both varieties, respectively (Fig. 1). Of the clinical C. neoformans var. grubii strains, 13 (54.2%) were genotype AFLP1 and 9 (37.5%) were AFLP1B; 2 (8.3%) were C. neoformans var. neoformans. Three (37.5%) strains out of the 8 (25%) environmental samples fell within the genotype AFLP1-cluster, 4 (50%) in the genotype AFLP1B-cluster and 1 (12.5%) in the hybrid genotype AFLP3-cluster (Fig. 1; Table 1).
Distribution of Cryptococcus neoformans mating-types, serotypes and genotypes.
| Serotype | Mating-type | AFLP genotype | ||||||
|---|---|---|---|---|---|---|---|---|
| Isolates | Serotype A | Non-typeable | α | Non-typeable | AFLP1 | AFLP1B | AFLP2 | AFLP3 |
| Clinical | 22 (91.7%) | 2 (8.3%) | 22 (91.7%) | 2 (8.3%) | 13 (54.2%) | 9 (37.5%) | 2 (8.3%) | – |
| Environmental | 5 (62.5%) | 3 (37.5%) | 5 (62.5%) | 3 (37.5%) | 3 (37.5%) | 4 (50%) | – | 1 (12.5%) |
| Total | 27 (84.4%) | 5 (15.6%) | 27 (84.4%) | 5 (15.6%) | 16 (50%) | 13 (40.6%) | 2 (6.3%) | 1 (3.1%) |
The antifungal susceptibility profiles for amphotericin B, fluconazole and voriconazole showed that all 32 C. neoformans strains were sensitive to these antifungal compounds. Table 2 depicts the average of the growth inhibition zones and its distribution.
DiscussionThe present study was performed in order to investigate the epidemiology of C. neoformans in a retrospective cohort of patients and from available environmental strains from Lima, Peru, using mating-type, serotype and genotype determination and antifungal susceptibility. By using AFLP genotyping, 16 (50%) of the strains were found to be genotype AFLP1 and 13 (40.6%) were genotype AFLP1B (representing C. neoformans var. grubii). Two strains (6.3%) were found to be genotype AFLP2, representing C. neoformans var. neoformans, and 1 (3.1%) was found to be genotype AFLP3 representing the hybrid between both varieties.
A previous large Ibero-Latin American epidemiological survey27 showed that from the 13 clinical strains from Lima, 12 (93.3%) were genotype AFLP1 and 1 (7.7%) was AFLP4 (C. gattii). The same study showed that the majority of the studied Latin American Cryptococcus strains fell within genotype AFLP1. In Amazon-Brazil, 31 (77.5%) isolates from a total of 40 strains were identified as AFLP1 and 9 (22.5%) as genotype AFLP6 (C. gattii); similarly, in Goiania-Brazil, 120 (97%) of the studied isolates were AFLP1 and 4 (3%) were found to be genotype AFLP5 (C. gattii).11,29
In Europe there was also a predominance of genotype AFLP1; however, a considerable percentage of clinical strains belong to genotype AFLP2 and AFLP3. In Madrid, Spain genotype AFLP1 represented 34 (58.9%) of the tested clinical strains.13 In Croatia, genotype AFLP1 represented 6 (40%) of the studied strains,28 while genotype AFLP2 strains represented 11 (19%) of the studied strains in Madrid and 6 (40%) in Croatia.13,28 The percentage of genotype AFLP3 was found to be 21 (32.2%) in Madrid and 3 (20%) in Croatia.13,28 A large study performed on 300 clinical strains from the Netherlands showed that genotype AFLP1 represented 245 (81.7%) of these strains, 36 (12%) genotype AFLP2, 14 (4.7%) genotype AFLP3, 1 (0.3%) genotype AFLP4, 1 (0.3%) AFLP6 and 3 (1%) genotype AFLP8.18
Unlike what was found in Europe, the distribution of C. neoformans genotypes in Taiwan25 is similar to what was observed in the current study, with 98 (98%) of genotype AFLP1 strains, 1 (1%) of genotype AFLP1A, and 1 (1%) AFLP4 (C. gattii).
Regarding antifungal susceptibility, it was observed that all the strains (clinical and environmental) were sensitive to amphotericin B, fluconazole and voriconazole. An epidemiological study in Cuba21 showed an antifungal sensitivity with low MICs against voriconazole and fluconazole, especially for voriconazole (MIC90 of 0.064μg/ml). A set of 120 investigated clinical and environmental samples from Goiana (Brazil) were found to be susceptible to amphotericin B, fluconazole, itraconazole and voriconazole.29
In India, the antifungal susceptibility was investigated for 308 clinical and environmental strains of C. neoformans (AFLP1; n=246, 80%) and C. gattii (AFLP4; n=62, 20%), which showed that 306 (99.4%) of the strains were sensible to the antifungal panel (amphotericin B, flucytosine, fluconazole, itraconazole and voriconazole), and only two clinical strains of C. neoformans var. grubii were found to be resistant to flucytosine (MIC>64μg/ml). Nevertheless, the strains of C. gattii were less sensible than the strains of C. neoformans.7
In 67 clinical strains isolated from HIV-infected patients in Kenya, 63 (94%) strains of C. neoformans and 4 (6%) of C. gattii were all susceptible to amphotericin B, ravuconazole and voriconazole, and 65 (97%) to fluconazole. The remaining 2 (3%) showed dose-dependent susceptibility to fluconazole.26
All these studies show that C. neoformans and C. gattii are susceptible to all the tested antifungals. These results are similar to those found in the current study.
There are very few studies in Peru on the epidemiology of C. neoformans and C. gattii. As mentioned above, an Iberoamerican study showed that the majority of the South American clinical strains belong to genotype AFLP1.27 Another study investigated the presence of C. gattii strains among 18 clinical strains from the culture collection of the National Institute of Health from Lima, Peru, by a canavanine-glycine-bromothymol blue test. However, it was determined that all strains belonged to C. neoformans.5 Another study investigated the presence of C. neoformans and C. gattii in the environment by sampling pigeon droppings in the city of Ica, Peru, but this study did not provide a differentiation between the two pathogenic Cryptococcus species.9
Also in Peru clinical studies have been conducted in patients with cryptococcal meningitis. A study performed at the Hospital Nacional Dos de Mayo in Lima showed that the combination of amphotericin B and fluconazole leads to an acceptable rate of sterilization of the cerebrospinal fluid after 10 weeks of treatment.10 A study performed in the Hospital Cayetano Heredia in Lima showed that a low Glasgow score and a serum antigen titer greater than 1024 were associated with higher mortality.6 Both studies did not differentiate between the two pathogenic Cryptococcus species. Current evidence from Peru suggests that the majority of clinical cases are due to C. neoformans var. grubii. However, there are clinical reports of the presence of C. gattii in Peru.4,14 One of the two Peruvian C. gattii strains (27) was recently found to be an interspecies hybrid between C. neoformans var. neoformans (AFLP2) and C. gattii (AFLP4).17
One limitation of our study is the small number of clinical samples. We have only analyzed the strains stored in our hospital culture collection. However, the study can be seen as a starting point for a national surveillance and research network. It will be interesting to carry out a nationwide prospective study to determine the presence of C. neoformans and C. gattii in clinical samples. Moreover, the environmental samples in our collection were obtained from pigeons, there were no samples obtained from soil or trees although it is known that both species can be found in these sources. Therefore, it is possible that the isolation of C. neoformans was favored above that of C. gattii. Likewise, due to the climatic conditions of Lima with low annual rainfall and a mild desert climate15 it was unlikely that the isolation of C. gattii in environmental samples was favored. Despite of the absence of other studies presenting a larger number of samples, we believe this work could serve as a reference for future prospective studies.
According to previous South American epidemiological studies, we noted that C. neoformans var. grubii was always the major isolated species. These studies were carried out in tropical climates, mainly in Brazil and Colombia, which may explain why the isolation of C. gattii was prevalent. Lima is considered a desert, with climatic features different than the major studied Latin American countries.
An interesting result was the finding of two strains of C. neoformans var. neoformans. One reason that could explain the latter is that these clinical samples came from El Callao, the main port in Peru. Moreover, our main international airport is located in the same area. The foreign traffic is heavy in this area, which could explain the presence of this genotype that is known to be more prevalent in Europe.
In conclusion, we observed that C. neoformans var. grubii (genotype AFLP1 and AFLP1B) and C. neoformans var. neoformans (genotype AFLP2) were the only cryptococcal species involved in human infections in Lima, Peru. All strains were susceptible to the antifungal compounds amphotericin B, fluconazole and voriconazole, which is consistent with previous studies.
Conflict of interestThe authors declare no conflict of interest.
Mercedes Tello is supported by a scholarship for postgraduate studies in Cellular and Molecular Biology, from the Consejo Nacional de Ciencia y Tecnología (CONCYTEC-Perú). Ericson Gutierrez is supported by a scholarship for postgraduate studies in Demography and Population, from the Belgian Development Cooperation CTB-Perú (L11PER016).