Histoplasmosis, caused by the dimorphic fungus Histoplasma capsulatum, represents an important public health problem, especially in urban environments where bats and humans cohabit indoors.
AimsTo detect the presence of H. capsulatum indoors, using samples of bat droppings collected in roost sites inside houses.
MethodsA Real-Time TaqMan PCR assay targeting the ITS1 region of the ribosomal DNA of H. capsulatum was carried out.
ResultsFifty-nine sampling points in the municipality of São Paulo were inspected, all of them located at inhabited places. H. capsulatum was isolated from nine samples.
ConclusionsThe rapid identification and monitoring of sites where the fungus is present may contribute to make a more reliable database of H. capsulatum distribution.
La histoplasmosis, causada por el hongo dimorfo Histoplasma capsulatum, representa un importante problema de salud pública, especialmente en los entornos urbanos donde los murciélagos y los humanos conviven en los espacios interiores.
ObjetivosDetectar la presencia de H. capsulatum en interiores mediante muestras de excrementos de murciélagos recogidas en sitios de reposo de estos animales dentro de las casas.
MétodosSe llevó a cabo un ensayo de PCR TaqMan® en tiempo real dirigido a la región ITS1 del ADN ribosómico de H. capsulatum.
ResultadosSe muestrearon 59 puntos en el municipio de São Paulo, todos ubicados en lugares habitados. H. capsulatum se aisló en nueve de estos lugares.
ConclusionesUna rápida identificación y control de los sitios donde se encuentra H. capsulatum contribuiría a la creación de una base de datos más sólida en lo referente a la distribución de este hongo.
Histoplasmosis, caused by the dimorphic fungus Histoplasma capsulatum, is an important public health problem in Brazil as information about its epidemiology is scarce.5,7 The fungus grows in soil enriched with birds or bats excrements, and infects humans during activities of soil disturbance.10 Bats acquire the fungus through inhalation and can develop a systemic disease, releasing spores in their feces.4 Therefore, the exposure to bat excrements represents an important source of infection, especially in urban environments as these animals are adapted to roosting in roofs, ceilings and other man-made shelters.6,9 In order to address the issue of whether or not the fungus is present inside houses, fecal samples were collected in roost sites directly into clean containers avoiding cross contamination. On ceilings with easy access, great extension and different environmental conditions, such as exccess of humidity or heat, the samples were collected from different points according to these conditions. These sampling points were selected based on complaints through the System of Assistance to the Citizen of the municipality of São Paulo.
DNA extraction was performed using the PowerSoil® Isolation Kit (MO BIO, USA) following the manufacturer's instructions with some modifications. A Real-Time TaqMan PCR assay (TaqMan® Gene Expression Assay, Applied Biosystems™, South San Francisco, CA, USA) was carried out following the manufacturer's instructions, and targeting the ITS1 region of the ribosomal DNA of H. capsulatum. The primers HcITS-54F (5′-ACCCTTGTCTACCGGACCTGTT-3′) and HcITS-204R (5′-TTTTGACTGGATTATTATCGCTCTCA-3′), and the probe HcITS-155 (5′ FAM-CGGTGAACGATTGGCGTCTGAGC-QSY 3′) were used as described by Buitrago et al.2 The target sequence was amplified in a final reaction of 25μl containing 2.5μl DEPC treated-water (Applied Biosystems™), 12.5μl TaqMan® Environmental Master Mix 2.0 (Applied Biosystems™), 2.5μl probe at 2.5 μM (Applied Biosystems™), 2.5μl of each primer (0.6μM) (Applied Biosystems™) and 2.5μl of DNA. DNA extracted from the H. capsulatum isolate MIC 1408/14 – culture collection – Mycology Section-ZCD/SP was used as the positive control, and DEPC treated-water was the negative control. The concentrations of the probe and primers were optimized, and those with the highest normalized reporter (ΔRn) and threshold cycle (Ct) were used to prepare the standard curve generated with serial 10-fold dilutions, wherein a 97% efficiency was obtained. Horse blood was used as an exogenous control, with the β-actin gene being the external target.
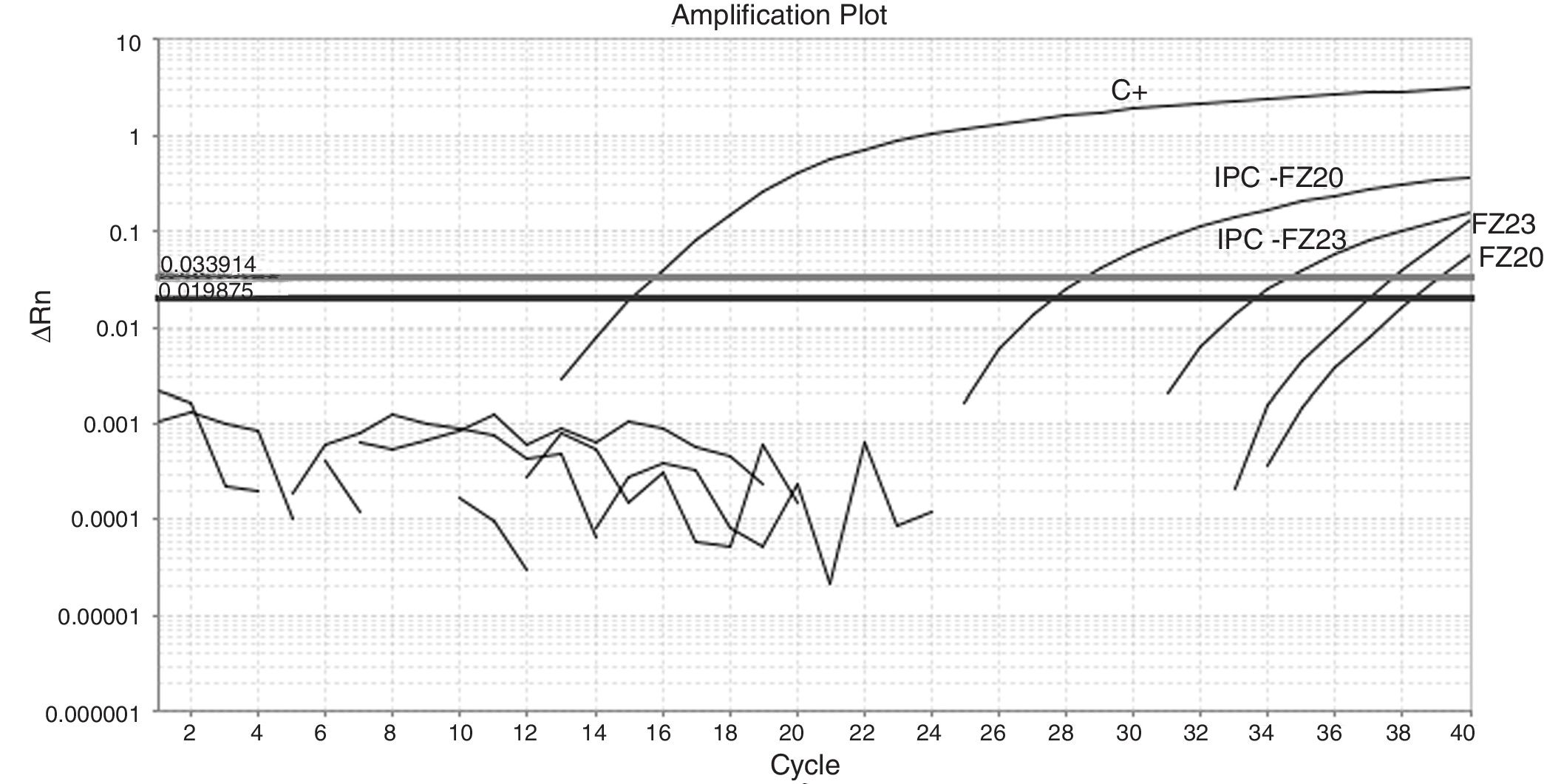
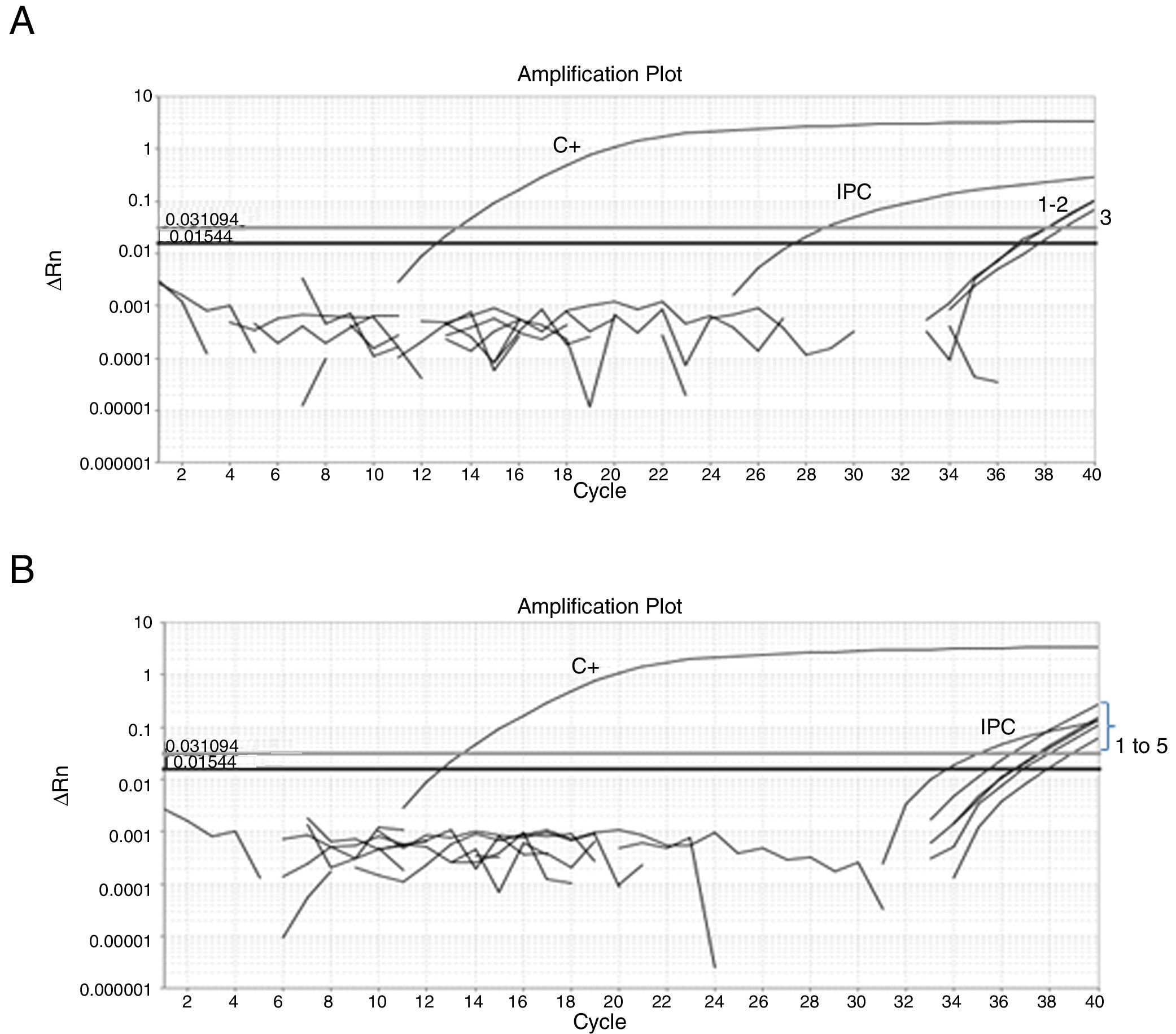
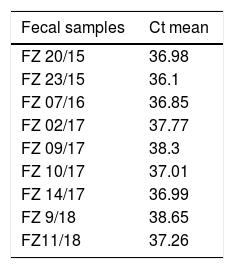
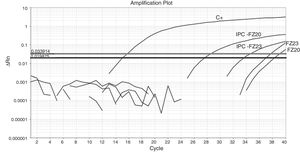
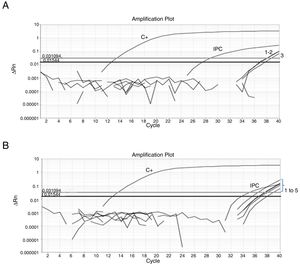
Fifty-nine sampling points were inspected, all of them located at inhabited places. H. capsulatum was isolated from 9 samples. The first positive samples, FZ20 and FZ23, showed Ct values of 38.26 and 37.1, respectively (Fig. 1). As the Ct obtained was higher than 37 cycles, the qPCR amplification products of both samples were submitted to electrophoresis in a 1.5% agarose gel, yielding a 180-bp band, as expected. The test was repeated in sextuplicate for the same samples (Fig. 2). The qPCR products were purified using QIAquick® PCR purification kit (Qiagen, Hilden, Germany) following manufacturer's instructions, and sent for sequencing to the Human Genome Research Centre at the University of São Paulo. DNA analysis was performed in the equipment ABI 3730 DNA Analyser (Applied Biosystems™), the sequencing reactions used BigDye™ Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems™) and the sequences were analyzed by Sequencing Analysis 5.3.1 software, using Base Caller KB. The sequences were compared to other sequences deposited at GenBank, showing 99% identity with H. capsulatum. The qPCR, electrophoresis and sequencing results obtained suggest that Ct values close to 38.5 indicate the presence of H. capsulatum, and that was the value used as cut-off. Table 1 shows those Ct values considered positive in the samples analyzed.
(A) Amplification plot of a qPCR reaction from a sextuplicate of the feces sample FZ20. Three replicates showed undetermined Ct and three showed the following Ct values: Ct1=36.9; Ct2=37.1 and Ct3=37.72; positive control (C+) Ct=12.6; exogenous internal positive control (IPC) Ct=28.75. (B) Amplification plot of a qPCR from a sextuplicate of the feces sample FZ23. One replicate showed undetermined Ct and five showed the following Ct values: Ct1=36.62; Ct2=35.47; Ct3=36.94; Ct4=36.56 and Ct5=37.91; positive control (C+) Ct=12.6; exogenous internal positive control (IPC) Ct=34.98; ΔRn: normalized reporter; Ct: threshold cycle.
The World Organization for Animal Health (OIE) highlights the need for detecting diseases that affect the wild animal population as a tool for a better understanding of the epidemiology of these diseases and their relation to the infection in humans.8 Yet this strategy is regrettably not implemented by public health systems in Brazil, as there is scarce reliable information to subsidize effective measures regarding the health surveillance of histoplasmosis. Additionally, the standard method for the detection of H. capsulatum is its isolation in culture. Although the latter being an easy method, the fungus is slow-growing, taking several days, even months, before a result is available. Moreover, feces are a heavily contaminated material, with the presence of fast-growing bacteria that can inhibit fungal growth, and there is a necessity of a confirmatory test after isolation.11 Due to these limitations, public health surveillance is impaired, as control actions regarding this disease cannot be taken in a timely manner. In this study, the use of a Real-Time TaqMan assay proved to be a very promising tool for histoplasmosis surveillance. It can also enable the improvement of control actions as infection sites are detected more rapidly, suggesting a potential reduction of clinical cases of histoplasmosis. Other studies demonstrated that some PCR protocols were successful when used to find the fungus in environmental sources,3,4,12 but they require a gel electrophoresis analysis, making the process more laborious and time-consuming. Real-time PCR for the detection of fungal DNA has been applied in many contexts and samples, especially for diagnostic purposes, and is specific and sensitive.1 This technique, compared to other PCR protocols, is more accurate and faster when applied to DNA extracted from clinical strains or culture extracts.2,11
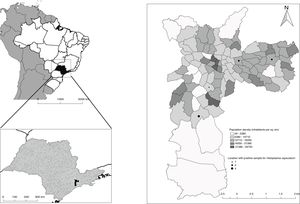
The sampling points where positive samples were identified corresponded to populated areas (Fig. 3). This finding is of great importance for epidemiological studies. The rapid identification and subsequent monitoring of sites within urban areas where the fungus was isolated contribute to draft a more reliable database of H. capsulatum distribution, as more samples can be processed in a shorter period of time, promoting a better understanding of histoplasmosis epidemiology.
This work was supported by the São Paulo Research Foundation (Fundação de Amparo à Pesquisa do Estado de São Paulo – FAPESP; Grant number 2014/06571-2).
Conflict of interestNone.
The authors thank Rosane Correa de Oliveira, from Zoonoses Surveillance Division, and Dr. Ana Paula de Arruda Geraldes Kataoka, Laboratory of Zoonoses and Vector-borne Diseases, for the institutional support and incentive to the development of the study. The authors also thank Dr. Gilda Maria B. del Negro, from Institute of Tropical Medicine – University of São Paulo-SP, and Dr. Mauro Cintra Giudice, from Oswaldo Cruz Colleges, for their technical support and expertise.