In Chile, the peony is the most important ornamental flower exported from the country. Gray mould is a phytopathological problem of this crop. This disease is caused by Botrytis cinerea and Botrytis paeoniae.
AimsWe carried out the first survey of Botrytis species associated with peony gray mould in Southern Chile to estimate the diversity of these pathogens.
MethodsDiseased peony leaves were collected from seven locations in Southern Chile covering a distance of 300km. The Botrytis isolates obtained were studied by morphological and molecular methods. Finally, a PCR assay using primers based on the necrosis and ethylene-inducing protein gene (nep1) was used to specifically identify B. paeoniae.
ResultsSeventeen isolates belonging to Botrytis genus were obtained, and all of them were pathogenic to peonies when inoculated in plants grown in a greenhouse. Morphological analyses showed that four isolates shared common characteristics, which distinguish them from the rest. Homology and phylogenetic analysis of G3PDH, as well as determination of the Bc-hch allele, allowed us to identify 12 isolates as B. cinerea, 4 as B. paeoniae and one isolate as Botrytis pseudocinerea. The PCR assay was found to be specific to B. paeoniae, amplifying a single band of 470bp.
ConclusionsThree Botrytis species involved in peony gray mould disease are present in Chile. This is the first time that both B. paeoniae and B. pseudocinerea have been reported to be present in the country and also that they affect peonies. Finally, to our knowledge, the PCR based method herein described is the first of its kind to be used to identify B. paeoniae.
La peonía es la principal flor ornamental de exportación en Chile. La pudrición gris, causada por los hongos Botrytis cinerea y Botrytis paeoniae, es uno de los problemas fitopatológicos más importantes de su cultivo.
ObjetivosRealizar una primera estimación de la diversidad de especies del género Botrytis asociadas a la pudrición gris de la peonía en el sur de Chile.
MétodosSe recogieron hojas de peonías con síntomas de pudrición gris en siete lugares del sur de Chile, cubriendo una distancia de 300km. Se estudiaron morfológica y molecularmente los aislamientos de Botrytis obtenidos. Finalmente, se desarrolló un ensayo de PCR específico para identificar B. paeoniae basado en el gen nep1.
ResultadosSe obtuvieron 17 aislamientos del género Botrytis; todos ellos causaron pudrición gris en plantas de peonía en ensayos de invernadero. Los análisis morfológicos mostraron que cuatro aislamientos compartían características comunes que los distinguen del resto. La homología y el análisis filogenético basado en el gen G3PDH y la determinación del alelo Bc-hch permitieron identificar 12 cepas como B. cinerea, 4 como B. paeoniae y un aislamiento como Botrytis pseudocinerea. Finalmente, el ensayo de PCR resultó específico para B. paeoniae al amplificar una sola banda de 470 pb.
ConclusionesTres especies de Botrytis están presentes en Chile asociadas a la pudrición gris de las peonías, y se describe por primera vez en este país la presencia de B. paeoniae y B. pseudocinerea como especies patógenas de los cultivos de esta planta. Hasta donde conocen los autores, se describe por primera vez un ensayo de PCR para identificar específicamente B. paeoniae.
Peony (Paeonia lactiflora Pall.) is an ornamental plant, native to China, and due to its high ornamental value it has become popular and has gained attention around the world.25 In Chile, peonies were introduced as a new option of economic activity in the south of the country leading this commercial crop to represent 46% of the total income value of all exported flowers.2
Gray mould is one of the most important diseases of the peony and it is caused by two fungal species: the non-host specialized plant pathogen Botrytis cinerea and the peony-specific pathogen Botrytis paeoniae.3 Both species have been reported to be widely distributed around the world such as in the USA,3 Iran16 and China,26 but the relative importance of these pathogens is still unknown. Despite the availability of taxonomic guides, there is a lack of alternative, efficient and reproducible methods for the identification of Botrytis species.9,16 However, several genes have been used to evaluate the phylogeny and evolution of Botrytis and could be used for molecular identification.20,21,24 In the case of B. cinerea, the presence or absence of two transposons, Boty and Flipper, in the genome of the pathogen allows for the establishment of the genotypes named vacuma (isolates without either transposon) and transposa (isolates with both transposons).8 Both genotypes are associated with differences in their ecology and population biology, such as in their host range and their genetic diversity, despite being frequently found in sympatry.4,8,15,17–19 Studies based on multiple gene genealogies have shown that B. cinerea is in fact a complex of at least two genetically different groups, Group I and Group II, which were proposed to be phylogenetic species.6,7 Group I isolates belong exclusively to the vacuma genotype and were formally described as a new species called B. pseudocinerea.24 Group II isolates include both vacuma and transposa genotypes and are referred to as B. cinerea.6,7
Due to the exportation increase of this species, peony cultivation has become more intensive, which has increased the risk of phytosanitary problems. Therefore, we conducted the first survey of its pathogens to characterize and to identify the species involved in the peony gray mould found in the main production area of Southern Chile. For this study, infected peony plants were collected from November to March during 2008 to 2010 in seven commercial fields of peonies located at seven sites covering a distance of 300km between Collipulli and Pilmaiquen (Table 1). Leaves with gray mould symptoms were incubated in a humid chamber until profuse sporulation was observed. Conidia were collected and plated onto half strength PDA plates (Potato-Dextrose Agar, Gibco/BRL, 1.5% agar) in order to obtain monosporic isolates.
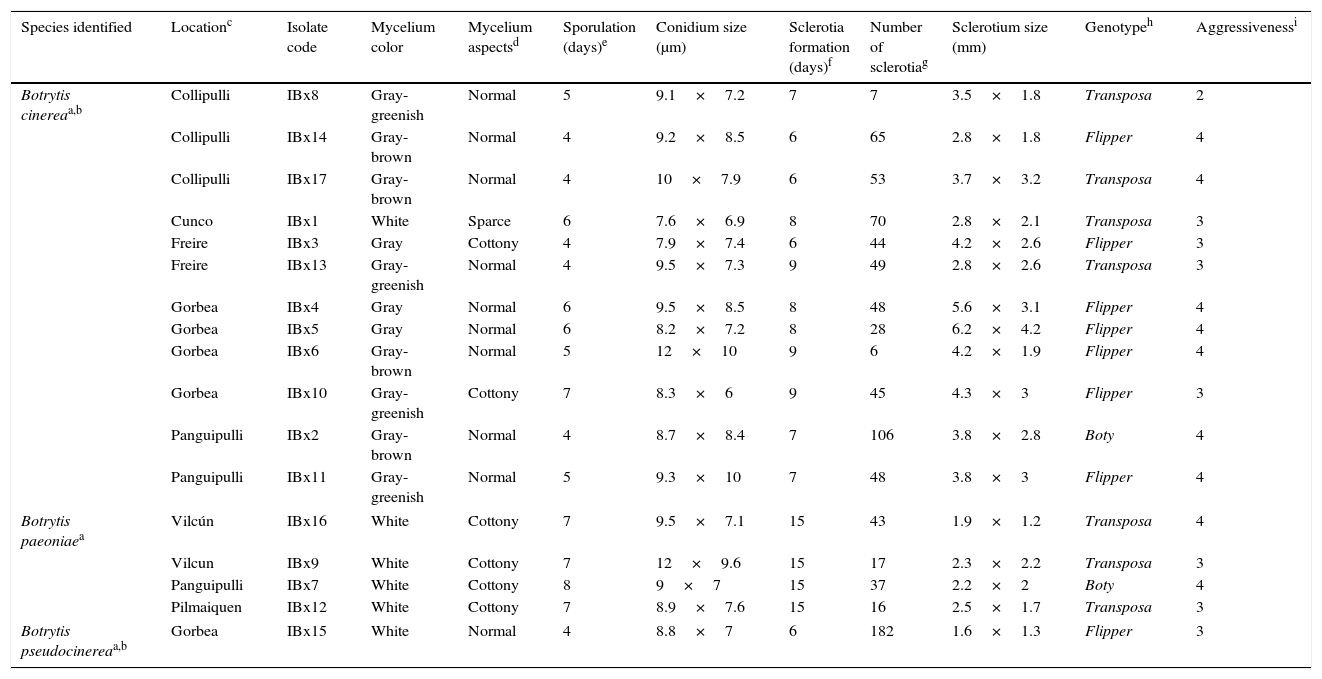
Origin, characterization and identification of Botrytis isolates collected from peonies in Chile.
| Species identified | Locationc | Isolate code | Mycelium color | Mycelium aspectsd | Sporulation (days)e | Conidium size (μm) | Sclerotia formation (days)f | Number of sclerotiag | Sclerotium size (mm) | Genotypeh | Aggressivenessi |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Botrytis cinereaa,b | Collipulli | IBx8 | Gray-greenish | Normal | 5 | 9.1×7.2 | 7 | 7 | 3.5×1.8 | Transposa | 2 |
| Collipulli | IBx14 | Gray-brown | Normal | 4 | 9.2×8.5 | 6 | 65 | 2.8×1.8 | Flipper | 4 | |
| Collipulli | IBx17 | Gray-brown | Normal | 4 | 10×7.9 | 6 | 53 | 3.7×3.2 | Transposa | 4 | |
| Cunco | IBx1 | White | Sparce | 6 | 7.6×6.9 | 8 | 70 | 2.8×2.1 | Transposa | 3 | |
| Freire | IBx3 | Gray | Cottony | 4 | 7.9×7.4 | 6 | 44 | 4.2×2.6 | Flipper | 3 | |
| Freire | IBx13 | Gray-greenish | Normal | 4 | 9.5×7.3 | 9 | 49 | 2.8×2.6 | Transposa | 3 | |
| Gorbea | IBx4 | Gray | Normal | 6 | 9.5×8.5 | 8 | 48 | 5.6×3.1 | Flipper | 4 | |
| Gorbea | IBx5 | Gray | Normal | 6 | 8.2×7.2 | 8 | 28 | 6.2×4.2 | Flipper | 4 | |
| Gorbea | IBx6 | Gray-brown | Normal | 5 | 12×10 | 9 | 6 | 4.2×1.9 | Flipper | 4 | |
| Gorbea | IBx10 | Gray-greenish | Cottony | 7 | 8.3×6 | 9 | 45 | 4.3×3 | Flipper | 3 | |
| Panguipulli | IBx2 | Gray-brown | Normal | 4 | 8.7×8.4 | 7 | 106 | 3.8×2.8 | Boty | 4 | |
| Panguipulli | IBx11 | Gray-greenish | Normal | 5 | 9.3×10 | 7 | 48 | 3.8×3 | Flipper | 4 | |
| Botrytis paeoniaea | Vilcún | IBx16 | White | Cottony | 7 | 9.5×7.1 | 15 | 43 | 1.9×1.2 | Transposa | 4 |
| Vilcun | IBx9 | White | Cottony | 7 | 12×9.6 | 15 | 17 | 2.3×2.2 | Transposa | 3 | |
| Panguipulli | IBx7 | White | Cottony | 8 | 9×7 | 15 | 37 | 2.2×2 | Boty | 4 | |
| Pilmaiquen | IBx12 | White | Cottony | 7 | 8.9×7.6 | 15 | 16 | 2.5×1.7 | Transposa | 3 | |
| Botrytis pseudocinereaa,b | Gorbea | IBx15 | White | Normal | 4 | 8.8×7 | 6 | 182 | 1.6×1.3 | Flipper | 3 |
Mycelium was considered as “normal” when white, gray or greenish aerial hyphae, sporulating or sclerotium were observed; “sparse” when a thin layer of mycelium stacked on the agar surface were observed, and “cottony” when abundant and dense white aerial mycelium was observed.
To characterize the isolates morphologically, mycelium was inoculated in the center of half strength PDA plates and was cultivated at 25°C in darkness. Observations of three replicate plates per isolate were made daily considering mycelium color and texture, the beginning of sporulation (determined as the time at which it was possible to observe the first conidiophores with conidia under the stereo microscope), the onset of sclerotia formation (considered as the time at which it was possible to detect the initial stage of sclerotium, as white or gray hyphae aggregated into small knots), and conidia and sclerotia size (determined by recording length and width of 50 conidia and 20 sclerotia under a light microscope or a stereo microscope using a micrometer installed in one of the ocular lenses).
Pathogenic assays were carried out in a greenhouse using 3-month-old peony cv. “Boule de Neige” plants potted in natural fertile soil. Each isolate was inoculated into two peony plants. Three leaves per plant were punctured and inoculated with a 4mm diameter PDA disk containing conidia and mycelium of the isolate. The fourth leaf (control) was inoculated with a PDA disk without the fungus. Plants were immediately sprayed with sterile distilled water and then were covered with a transparent plastic bag for 72h and kept in a greenhouse at 22–26°C for nine days. Then, the diameter of the necrotic lesion formed around each PDA disk was measured. Aggressiveness was determined using a scale of 0–4 defined as: 0, absence of lesion; 1, less than 2mm lesion; 2, 2–3mm lesion; 3, 3–5mm lesion; and 4, over 5mm lesions.
All of the 17 isolates selected for this study produced the characteristic gray mould lesion in the peony leaves under greenhouse conditions (Table 1), demonstrating all of them were pathogenic to the host plant. The level of infection was high and fairly homogeneous among the isolates, most of them scoring an aggressiveness of 3–4. Morphologically, at least two groups were detected when cultivated on PDA plates (Table 1). One group composed of isolates IBx7, IBx9, IBx12 and IBx16 showed fairly similar phenotypes such as abundant white aerial mycelium, conidia produced after seven days, and sclerotia observed after 15 days. The remaining isolates presented a mycelium such as that observed in Botrytis species with sporulation developing after 4–7 days and sclerotia being produced after 6–9 days.
The identification of Botrytis species was done by sequencing and phylogenetic analysis. In this regard, genomic DNA was extracted from each isolate as previously described,18 and a fragment of the G3PDH gene was amplified by PCR20 and sequenced at Unidad de Secuenciamiento Automático de ADN (Pontificia Universidad Católica de Chile). The obtained sequences (GenBank accession numbers KF015582 to KF015598, Fig. 1) were first analyzed for homology to known sequences with the BLAST program and the nucleotide databases.1 All sequences obtained showed corresponding matches to B. cinerea or B. paeoniae except for the isolate IB×15 that was homologous to B. pseudocinerea (JN692414). For the phylogenetic analysis, the corresponding G3PDH gene fragment of B. cinerea (AJ705005, AJ705006, AM231156 and AM231160), B. paeoniae (AJ705027, AJ705028, EF36130 and EF677131) and other Botrytis species (B. tulipae AJ705042; B. gladiorum AJ705020; B. squamosa AJ705038 and B. elliptica AM231166) were considered. Sclerotinia sclerotiorum (AF417110) and Monilinia fructigena (AJ705043) were used as out-group controls. For the phylogenetic analysis, the sequences were first aligned and then analyzed using the neighbor-joining method of the distance matrix with 1000 bootstrap replicates using the MEGA4.0 program.22 The dendrogram obtained (Fig. 1) showed that 12 isolates (IBx1, IBx2, IBx3, IBx4 IBx5 IBx6 IBx8 IBx10 IBx11 IBx13 IBx14 and IBx17) clustered with B. cinerea and four isolates (IBx7, IBx9, Bx12 and IBx16) formed a group with B. paeoniae. The species B. cinerea and B. pseudocinerea were confirmed by determination of the Bc-hch allele as previously described.7
Phylogenetic dendrogram constructed using neighbor-joining analysis of a GPDH fragments. The fungal species considered in this analysis and their corresponding GenBank accession numbers are indicated on the figure. The dendrogram was rooted with Sclerotinia sclerotiorum and Monilinia fructigena. Bootstrap values are given at each branch. The bar indicates the expected changes per site.
Despite being expected, the presence of B. paeoniae has not been previously described in Chile. These isolates were collected in three different locations including Vilcún, Panguipulli and Pilmaiquen, thus suggesting that this pathogen is widely distributed throughout the country. Because peony production is a very recent economic activity in Southern Chile, and also due to the host specialization of this species, it is highly probable that B. paeoniae was introduced to the country along with the plant.
Detection of B. pseudocinerea was an unexpected finding and to our knowledge this is the first time this species has been reported in peonies and also in Chile. As previously described for other crops in the rest of the world,5,10,11,13–15,19,24B. pseudocinerea seems to be infrequent in peonies in Chile. However, a more detailed study is needed to establish the real frequency of this species.
In order to make comparisons with Botrytis populations previously described in Chile, isolates were genotyped using PCR to detect the presence of Boty and Flipper transposons as previously described.17 Noteworthily, we found that the isolate IBx15 contained only the Flipper transposon (Table 1). In France, B. pseudocinerea populations have been described to be composed of only vacuma isolates.6,7,24 Our result agrees with those reported in Hungary5 and New Zealand,11 where a wide diversity of transposon types have been found in the B. cinerea populations. In addition, a high frequency of the Flipper genotype (7 out of 12 isolates) was found in the isolates identified as B. cinerea, while the other genotypes were mainly transposa (4 out of 12 isolates) and only one isolate was identified as Boty (Table 1). The Flipper genotype has been nearly absent in most commercial crops studied4,10,13,15,17 and has only been detected at significant levels in six plant hosts in Greece19 and in vineyards in Hungary.23 The particular distribution of these genotypes in the B. cinerea isolates suggests that the population of pathogens present in peonies might be different from those previously described in other crops in Chile.4,17,18
Finally, a specific PCR-based assay was established based on the necrosis and ethylene-inducing protein 1 gene (nep1).21Nep1 sequences corresponding to B. paeoniae (AM087032 and AM087033) were aligned with those corresponding to B. cinerea (AM087032, DQ211824 and JN672679) using the Clustal Omega program [http://www.ebi.ac.uk/Tools/msa/clustalo/help]. Primers BpNP1-F41 (5′-GCTACAGTCCCTCACGATTCG-3′) and BpNP1-R510 (5′-TAAGGTGGGGTTGGGGAGGG-3′) were designed using the Fast-PCR program.12 DNA amplifications were done with GoTaq Hot Start Polymerase (Promega) under reaction conditions recommended by the manufacturer; primers were used at 200nM, DNA ranged from 5 to 20ng, and the final volume of the reaction was 20μl. The program cycles were 2min at 95°C followed by 35 cycles of 20s at 95°C, 20s at 58°C and 20s at 72°C, and a final extension step of 5min at 72°C. Reactions were carried out in My Cycler™ thermal cycler (BioRad). Amplified products were visualized on 1% agarose gels stained with SafeView nucleic acid stain (NBS Biologicals Ltd), illuminated under UV light and digitally recorded. Other Botrytis isolates included in these assays were Botrytis aclada 961, Botrytis allii 403, Botrytis byssoidea 94, Botrytis squamosa 1107, Botrytis tulipae 9701 and B. paeoniae strains 771 and 16084 (a donation from Jan Van Kan, Wageningen University). Our results showed that the designed primers amplified a single DNA band of the expected size specific to B. paeoniae isolates (470bp, Fig. 2).
PCR identification of B. paeoniae using BpNP1-F41 and BpNP1-R510 primers. Lanes 1–5 correspond to the DNA samples of B. aclada, B. allii, B. byssoidea, B. squamosa and B. tulipae. Lanes 6–10 correspond to the DNA of B. paeoniae, isolates 1908, IBx7, IBx9, IBx12 and IBx16. Lane 11 corresponds to isolate IBx15 of B. pseudocinerea. Lanes 12–15 correspond to DNA samples of B. cinerea isolates IBx1, IBx2, IBx6 and IBx16. Lane 16 represents the 1kb ladder DNA.
In conclusion, B. cinerea, B. paeoniae and B. pseudocinerea, are present in Chile and are involved in peony gray mould disease. This study represents the first report of B. paeoniae and B. pseudocinerea existence and also that they affect peonies in this country. Finally, to our knowledge, the PCR-based method herein described is used for the first time to identify B. paeoniae.
Disclosure statementAuthors have nothing to declare.
Conflict of interestThe authors declare that they have no conflict of interest.
Authors kindly thank Dr. Jan Van Kan (Laboratory of Phytopathology, Wageningen University, Wageningen, the Netherlands) for kindly providing the DNA samples included in this study. This work was partially supported by Fundación para la Innovación Agraria, project PIT-2007-0247 granted to G. Chahin.