Candiduria is a common infection among hospitalised patients. Although the clinical relevance of yeasts in urine is not clearly defined, fungal urinary tract infections have increased significantly in the last decades, becoming a growing public health problem. Candida albicans is the most commonly reported species in urinary infections, although other species of the genus are becoming particularly important, because some of them are linked with resistance to antifungal drugs.
AimsThis study aimed to evaluate the frequency of Candida species causing candiduria in a hospital in Honduras.
MethodsA simple and cost-effective PCR-RFLP approach was used, by amplifying a partial sequence of the ribosomal ITS1-ITS2 region and a subsequent digestion with the enzyme MspI.
ResultsDuring 2016, an analysis was performed on 73 urine samples from patients of different ages. Seven species were found. Candida albicans/dubliniensis was the most frequent species (30%); Candida glabrata (28.8%) was the most isolated among the rest of the species. Candida kefyr was the least frequent species found (2.5%).
ConclusionsThis study shows, for the first time in Honduras, the frequency of the Candida species isolated from urine using PCR-RFLP for their identification. This approach could be applied in future epidemiological studies at local and national level.
La candiduria es una infección común entre pacientes hospitalizados. Aunque la relevancia clínica de las levaduras en orina aun no está del todo esclarecida, el número de infecciones urinarias por hongos se ha incrementado en las últimas décadas, convirtiéndose en un creciente problema de salud pública. Candida albicans es la levadura más común en las infecciones urinarias, si bien otras especies del género están adquiriendo importancia debido a la resistencia a las drogas antifúngicas asociada a algunas especies.
ObjetivosEste estudio pretendió evaluar la frecuencia de especies de Candida responsables de candiduria en un hospital nacional de Honduras.
MétodosSe utilizó un análisis in silico y PCR-RFLP de una región parcial de la secuencia ribosomal ITS1-ITS2, con posterior digestión mediante la enzima MspI.
ResultadosSe analizaron 73 muestras de orina de pacientes de diferentes edades durante el año 2016. Se encontraron siete especies diferentes. Candida albicans/dubliniensis fue la especie más común (30%); Candida glabrata (28,8%) fue la más frecuente entre el resto de especies. Candida kefyr fue la especie menos encontrada (2,5%).
ConclusionesEste estudio muestra la frecuencia de especies de Candida aisladas a partir de orina, e identificadas mediante la técnica PCR-RFLP por primera vez en Honduras. Este enfoque podría ser aplicado para futuros estudios epidemiológicos a nivel local y nacional.
Candida species are the most commonly isolated fungi in urine cultures, especially among hospitalized patients.3,15Candida urinary infections are a growing health problem for immunosuppressed and debilitated patients, or those receiving long-term antibiotic therapy.23 Although the clinical relevance of yeasts in urine is not clearly established, many authors agree that counts greater than 103–104CFU/ml suggest primary or disseminated infection,24 rather than simple contamination or colonization.
The most common etiological agent of candiduria is Candida albicans1; however urinary tract infections caused by other species of the genus have increased significantly in recent years.12,19 Some of the most common species causing candiduria are Candida glabrata, Candida tropicalis, Candida parapsilosis, Candida krusei and Candida kefyr.1,25 This fact raises special concern because of the low susceptibility to antifungal drugs showed by some Candida species, which may lead to difficulties or failures in the treatment.5
The identification of yeast species causing candiduria and the evaluation of their susceptibility to antifungal drugs are a major problem for clinical laboratories in low-income countries due to financial constraints. Some of these laboratories are restricted, due to the lack of resources, to only report the number of CFU/ml in urine; VITEK® 2 YST ID card, API Candida, API 20C, API ID32C, or CHROMagar Candida Medium are expensive methods when there is a lack of resources. For this reason, in Honduras, the information regarding Candida species causing urinary infections in public hospitals is scarce. Implementing a less costly but reliable technique for the identification of Candida species could help to fill this knowledge gap. Several alternative strategies have been proposed,13,29 however most of them require expensive equipment and a higher level of technical expertise. This study aimed to determine the frequency of species causing candiduria in hospitalized patients attending a public hospital in Honduras through a very simple molecular approach17 in order to assess the frequency of infections and the possibility of its further implementation as a routine test.
Materials and methodsPopulation and culture methodA total of 73 yeast cultures from urine samples were analyzed. The samples were obtained from patients of all ages hospitalized in a national facility in Tegucigalpa, Honduras, between July and September 2016. Only urine samples revealing yeasts in the microscopic analysis were cultured by calibrated loop (0.001ml) onto Sabouraud agar supplemented with 80μg/ml of gentamicin. Cultures were incubated at 37°C and read within 24h. The number of cultivable yeasts was established by the CFU counts.
DNA extractionA DNA extraction protocol using organic solvents was performed in the isolated yeast colonies. Briefly, an initial cell lysis step was carried out from one loop of an axenic culture suspended in 1000μl of buffer composed of 10mM Tris, pH 8; 1mM EDTA, pH 8; and 100mM NaCl. This suspension was heated at 100°C for 1min and then stirred for 1min in a micro-MiniBeadBeater® system (Bio Spec products Inc.) with 0.1mm glass beads. Four hundred μl were recovered and one volume of phenol – chloroform (1:1) was added, mixed and centrifuged at 10,000rpm. The aqueous phase was transferred to a new vial and precipitated with one volume of ice cold isopropanol and 1/10 volume of sodium acetate (3M, pH 5.2). Three washes in 70% ethanol were performed. The dried pellet was resuspended in 50μl of nuclease-free water. The DNA concentration was calculated with a NanoDrop spectrophotometer (Thermo Fisher Scientific Inc.), diluted to a final concentration of 40ng/μl and stored to −20°C until further use.
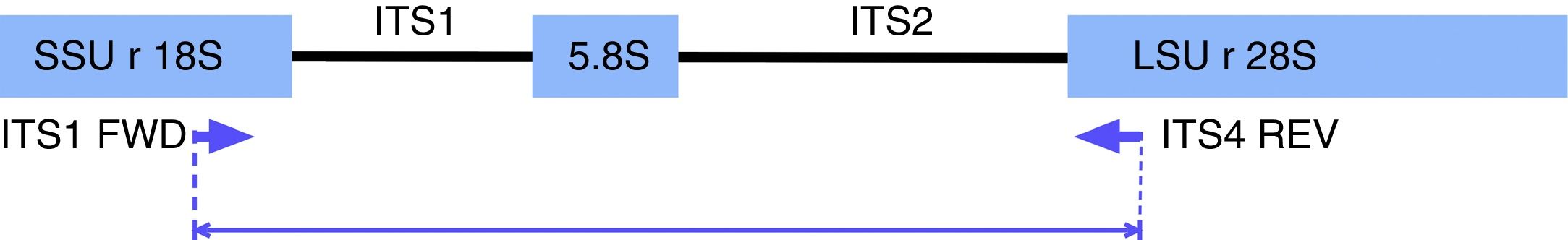
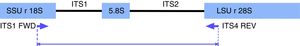
PCR-RFLP for yeast species identificationPCR reactions were performed according to Mirhendi et al.17,28 with modifications. Briefly, amplifications were carried out under the following conditions in a 50μl volume: 25μl of PCR Master Mix (Promega Corp. Madison, WI, USA), 1μl of 10μM ITS1 and ITS4 primers (Fig. 1): 5′-TCCGTAGGTGAACCTGCGG-3/5′-TCCTCCGCTTATTGATATGC-3′, and 1μl of DNA (40ng/μl). Reactions were carried out with an initial denaturation step at 95°C for 5min, 37 cycles of 95°C for 30s, 56°C for 30s, and 72°C for 30s, with a final extension at 72°C for 5min. Amplicons were visualized in 1.5% agarose gel electrophoresis with ethidium bromide. Ten microliters of PCR products were digested for 2h at 37°C with 2μl of buffer, 0.2μl of 10μg/μl acetylated BSA, and 0.5μl of the restriction enzyme MspI (10U/μl) (Promega Corp.). The restriction fragment pattern was analyzed on 2% agarose gel in 0.5X Tris-Acetate-EDTA (TAE) buffer and visualized in a BioDoc-It Imaging System (UVP, LLC; Upland, CA).
In silico analysisIn order to confirm the restriction patterns of previously reported Candida species17,28 as well as of other uncommon species, homologous sequences corresponding to the fungal ITS1-ITS2 region were imported from the BLAST tool of NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi) into the Geneious® 7.1.5 software (Biomatters Ltd, Auckland, New Zealand). Sequences were trimmed to include a partial fragment of the small subunit ribosomal RNA gene, the internal transcribed spacer 1, the 5.8 S ribosomal RNA gene, the internal transcribed spacer 2, and a partial sequence of the large subunit ribosomal RNA gene. Primers ITS1 and ITS4 flanked all sequences. The enzyme MspI was applied in the analysis to demonstrate putative cutting sites and restriction patterns for most of the available species (Fig. 2).
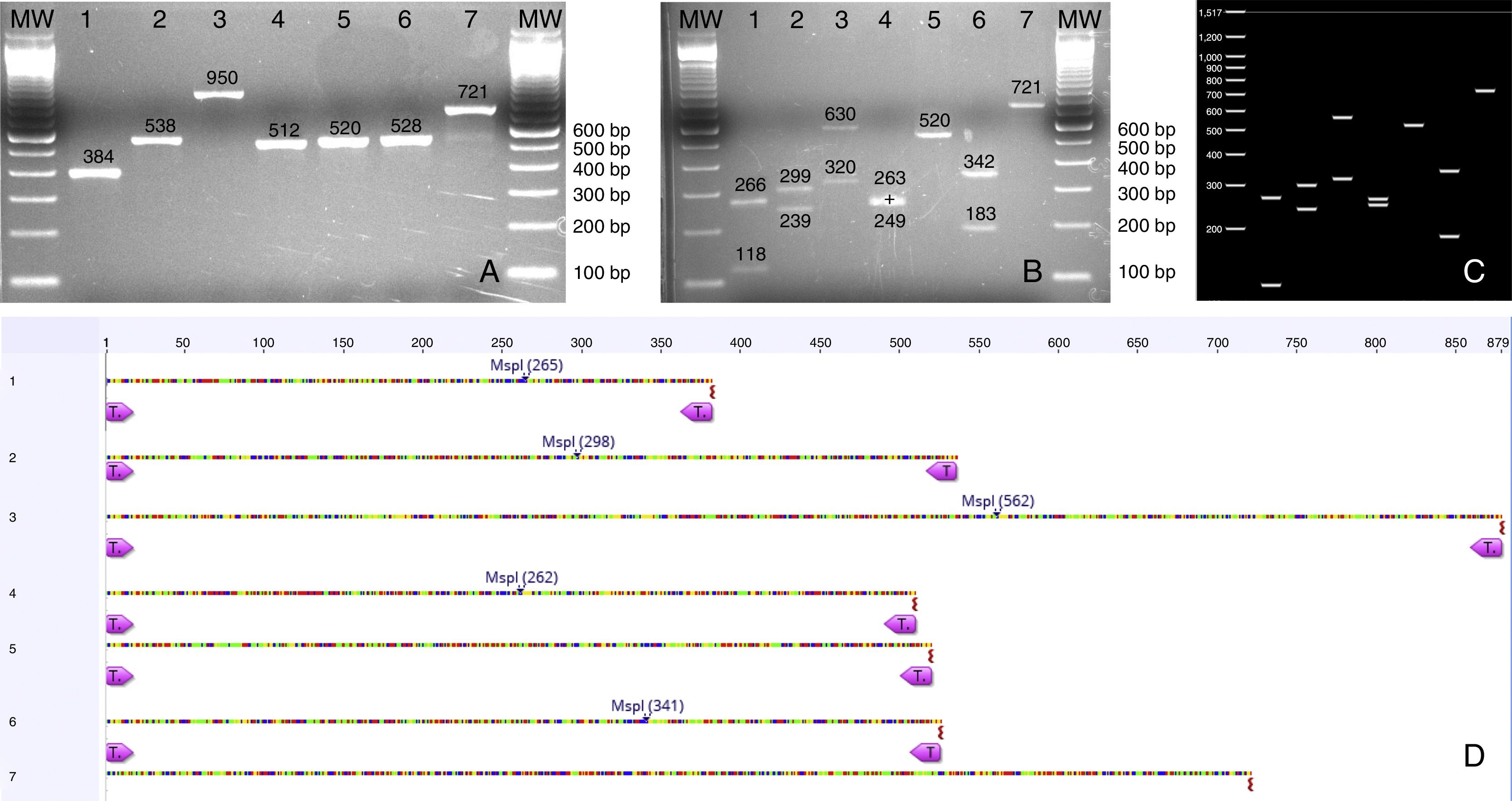
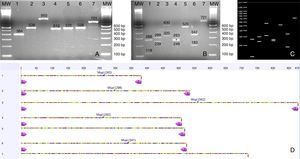
(A) PCR products from seven Candida species isolated from urine samples showing differential amplicon sizes. 1: C. lusitaniae; 2: C. albicans; 3: C. glabrata; 4: C. krusei; 5: Candida parapsilosis; 6: C. tropicalis; 7: C. kefyr; (B, D) In silico analysis of restriction profiles using MspI for the same seven yeast species; (C) Restriction results revealing complete coincidence with the in silico analysis.
To confirm the identity of the amplified products, seven gene fragments were selected according to molecular weight, and an ulterior sequencing was carried out on both strands with their respective ITS primers following standard protocols at Macrogen Corp. facilities (https://www.macrogenusa.com). Sequences were trimmed at both 5′ and 3′ ends with the Geneious® 7.1.5 software and queried against international databases contained in NCBI. The higher similarity index was recorded.
ResultsUrine culture and yeast countsSeventy-two (98.6%) urine cultures showed more than 103 CFUs, and one culture (1.36%) revealed less than 100 CFUs. Five cultures (6.84%) showed mixed candiduria and four of them (5.48%) reported two different yeast species, while the fifth culture (1.36%) revealed more than two yeast species. In none of the cases the same combination of species was reported.
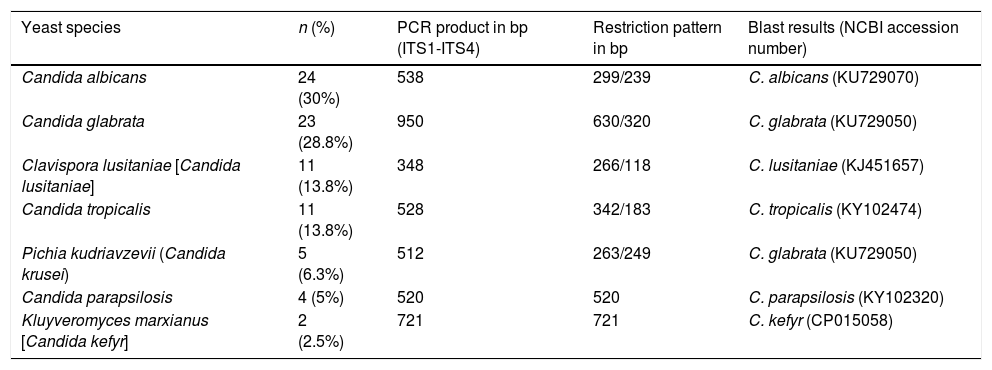
Yeast species identified through PCR-RFLPA total of 73 yeast colonies were analyzed and putatively identified through a PCR-RFLP approach using the ITS1-5.8 S-ITS2 ribosomal region. No phenotypic method for species identification was carried out. PCR products ranging from 512 to 950bp were obtained. The PCR-RFLP using MspI as restriction enzyme identified the following isolates: C. albicans/dubliniensis, C. tropicalis, C. glabrata, C. krusei, C. parapsilosis, Candida lusitaniae, and C. kefyr. The two most common species were C. albicans/dubliniensis and C. glabrata, with 30% and 28.8% of the isolates respectively (Table 1).
Yeast species identified through PCR-RFLP and sequencing. PCR product sizes, restriction profiles, and the most similar results from a BLAST search according to Max score, query cover and E-value, are shown.
| Yeast species | n (%) | PCR product in bp (ITS1-ITS4) | Restriction pattern in bp | Blast results (NCBI accession number) |
|---|---|---|---|---|
| Candida albicans | 24 (30%) | 538 | 299/239 | C. albicans (KU729070) |
| Candida glabrata | 23 (28.8%) | 950 | 630/320 | C. glabrata (KU729050) |
| Clavispora lusitaniae [Candida lusitaniae] | 11 (13.8%) | 348 | 266/118 | C. lusitaniae (KJ451657) |
| Candida tropicalis | 11 (13.8%) | 528 | 342/183 | C. tropicalis (KY102474) |
| Pichia kudriavzevii (Candida krusei) | 5 (6.3%) | 512 | 263/249 | C. glabrata (KU729050) |
| Candida parapsilosis | 4 (5%) | 520 | 520 | C. parapsilosis (KY102320) |
| Kluyveromyces marxianus [Candida kefyr] | 2 (2.5%) | 721 | 721 | C. kefyr (CP015058) |
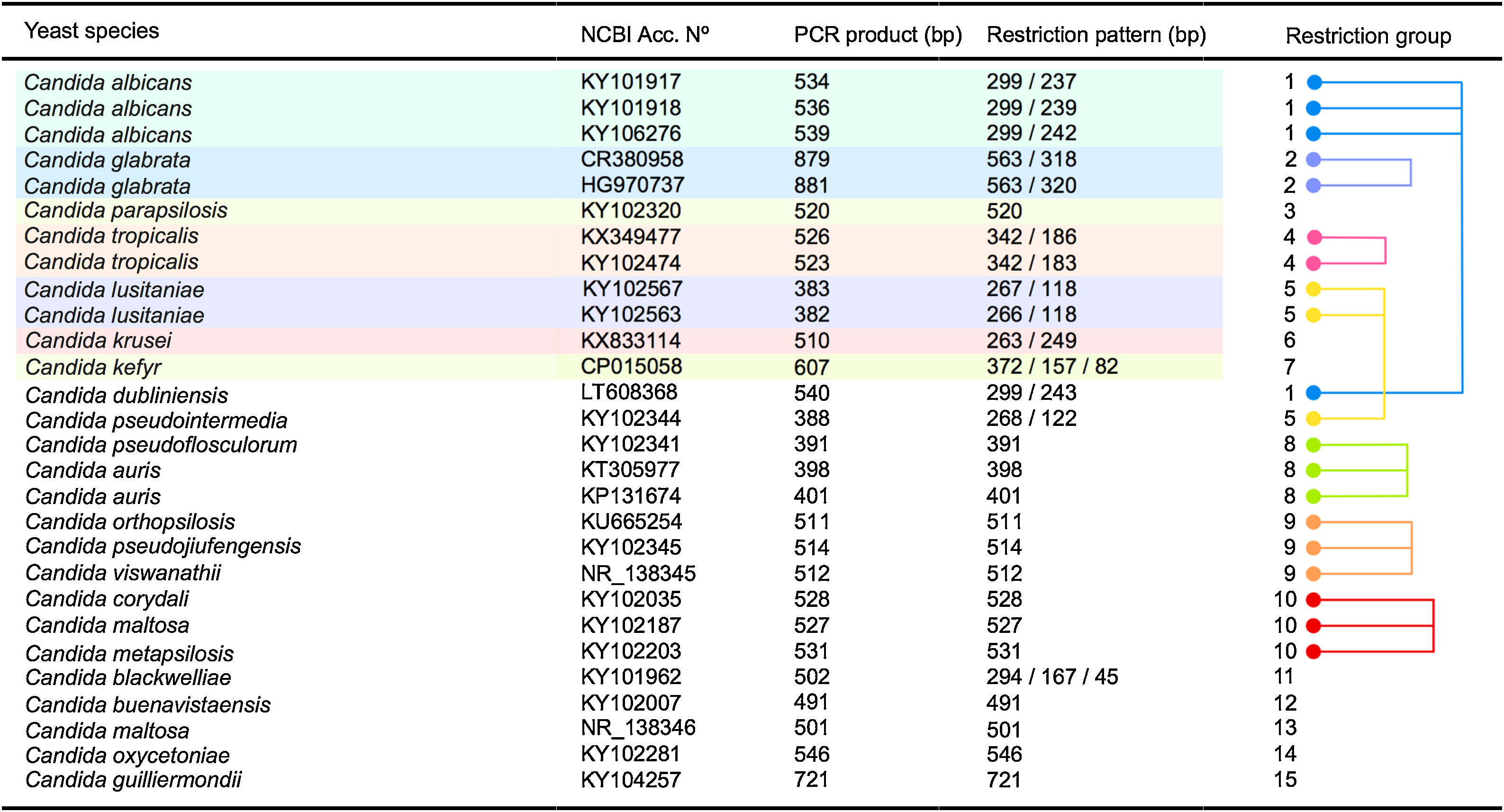
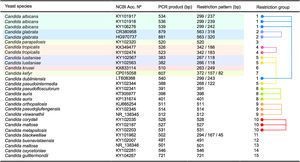
As shown in Fig. 3 an in silico analysis of sequences from 21 yeast species revealed fifteen different restriction groups. Ten species show a unique RFLP pattern (Candida blackwelliae, Candida buenavistaensis, C. glabrata, Candida maltosa, Candida oxycetoniae, C. parapsilosis, C. tropicalis, C. krusei, Candida guilliermondii and C. kefyr). The remaining 11 species shared a common restriction profile with a second or third individual as follows: Group 1 composed by C. albicans and C. dubliniensis; Group 10 (Candida corydali/C. maltosa/Candida metapsilosis); Group 9 (Candida orthopsilosis/Candida pseudojiufengensis/Candida viswanathii); Group 5 (C. lusitaniae and Candida pseudointermedia), and Group 8 (Candida pseudoflosculorum and Candida auris).
To demonstrate the reliability of the in silico analysis, more than one accession number for five species were included: C. albicans (n=3 accession numbers), C. glabrata (n=2), C. tropicalis (n=2), Clavispora lusitaniae (n=2), and C. auris (n=2). PCR product sizes before and after restriction were similar between the same species, except for Candida maltosa, which showed a PCR product with two different sizes for the two analyzed accession numbers (Fig. 3). This could be due to an erroneous identification of this particular accession or to a real polymorphism within the species.
In order to evaluate the capacity of the PCR-RFLP method to identify the isolated species, amplification products that showed a unique size were further sequenced. As shown in Table 1, all sequenced isolates were correctly identified at the species level, demonstrating a perfect coincidence between both methods.
DiscussionPresence of yeasts in urine may have different interpretations: (a) they could be contaminants with no etiological relevance in urinary tract infections14; (b) they could be the local consequence of a disseminated infection or candidemia,16 or (c) they could be actually the cause of a urinary infection, and in some cases even the focus of a systemic infection.8 In this study, 72 urine cultures with more than 103 CFUs were included. Although the definition of candiduria remains unclear, several authors agree on yeast counts ranging from 103 to 105CFUs/ml as clinically relevant.1,2,6,24 For this reason 72 out of 73 isolated microorganisms from urine samples were considered the primary cause of genitourinary infections rather than being only colonizing the mucosa. A mixed candiduria was found in five cultures (6.84%). This result is in agreement with several authors that report around 5% of urinary infections caused by more than one yeast species.2,15
Polymorphisms in both length and sequence are the main reason why the nuclear ribosomal internal transcribed spacer (ITS) region has been currently accepted as the universal DNA barcode marker for fungi, including the Ascomycetes yeasts.22 Herein, the 73 yeast colonies were putatively identified as seven different yeast species through a restriction analysis of the ITS ribosomal region. The two most common isolated species in this study were C. albicans/dubliniensis (30%) and C. glabrata (28.8%), followed by C. lusitaniae and C. tropicalis (13.8% each), and to a lesser extent C. krusei, C. parapsilosis and C. kefyr. All those seven species have been described as pathogens in urinary tract infections.20 A comprehensive review sets forth 50–70% of all urine isolates are C. albicans, followed by C. glabrata and C. tropicalis.1 A second review summarizes the most common Candida species in patients with candiduria from six different studies,4 where three of them report C. albicans and C. glabrata as first and second etiological agents respectively.10,15,24 In contrast, C. lusitaniae and C. kefyr have been reported less frequently as cause of candiduria.26
The RFLP analysis using the MspI enzyme on seven yeast species showed expected DNA profiles from ribosomal ITS fragments when compared with the in silico analysis (Fig. 2). In addition, we included in the in silico analysis sequences of less common yeast species causing human infections, for example the “–psilosis group”,7C. kefyr, as well as Candida species commonly associated with plants or insects,9,21 unlikely to cause human infections. C. auris sequences were also analyzed due to its importance as emerging yeast in invasive infections.27
Fifteen different restriction groups were obtained from the in silico approach, identifying 10 out of 21 species (Fig. 3). The remaining 11 species could not be completely identified through this approach. This apparent low specificity of the method could be considered a disadvantage for the correct identification of yeast species. However, this limit can be easily overcome when a second restriction enzyme is applied to differentiate between those species with a similar MspI-profile. For example, C. albicans and C. dubliniensis could be distinguished through an ulterior RFLP assay using the BlnI enzyme.18 Enzymes commonly used in the laboratory can separate the species within the other 4 restriction groups described herein. HindIII or HhaI could separate the species in the restriction Group 10 (including C. corydali/C. maltosa/C. metapsilosis); HaeIII differentiated the species in Group 9 (C. orthopsilosis/C. pseudojiufengensis/C. viswanathii); HhaI or HinfI separated the species in Group 5 (C. lusitaniae and C. pseudointermedia), and species in Group 8 (C. pseudoflosculorum and C. auris) are discriminated by AluI or TaqI. Confirmation through Sanger sequencing of seven isolates proved that a PCR-RFLP approach could be potentially useful for the identification of some of the most common yeast species causing candiduria, according to identifiable restriction profiles.
This method has also the advantage of being implementable in a hospital laboratory, and it is relatively inexpensive compared to commercial solutions based on phenotypic approaches, which are able to identify only the most common Candida species. Early identification of Candida species is also important for timely drug therapy and controlling the spread of virulent or resistant strains in a sanitary facility.17 In addition, more informative molecular markers such as multilocus sequence typing have proven to be a powerful tool to find out whether a patient has a recurrence with the same strain or not,11 or to demonstrate concordance between candidemia and candiduria,3 which could not be evidenced by more conventional approaches.
In conclusion, as far as we know, this is the first study to describe Candida species isolated from patients with candiduria in Honduras through a molecular approach, where C. albicans/dubliniensis and C. glabrata were the two most common species.
Conflict of interestsThe authors declare no conflict of interests.
We would like to thank the Research Directorate of the National Autonomous University of Honduras (DICYP-UNAH) for the financial support and to the Mycology Laboratory of the national Hospital for providing the urine samples.