Candida auris is unique due to its multidrug resistance and misidentification as Candida haemulonii by commercial systems. Its correct identification is important to avoid inappropriate treatments.
AimsTo develop a cheap method for differentiating C. auris from isolates identified as C. haemulonii by VITEK2.
MethodsFifteen C. auris isolates, six isolates each of C. haemulonii and Candida duobushaemulonii, and one isolate of Candida haemulonii var. vulnera were tested using CHROMagar Candida medium supplemented with Pal's agar for better differentiation.
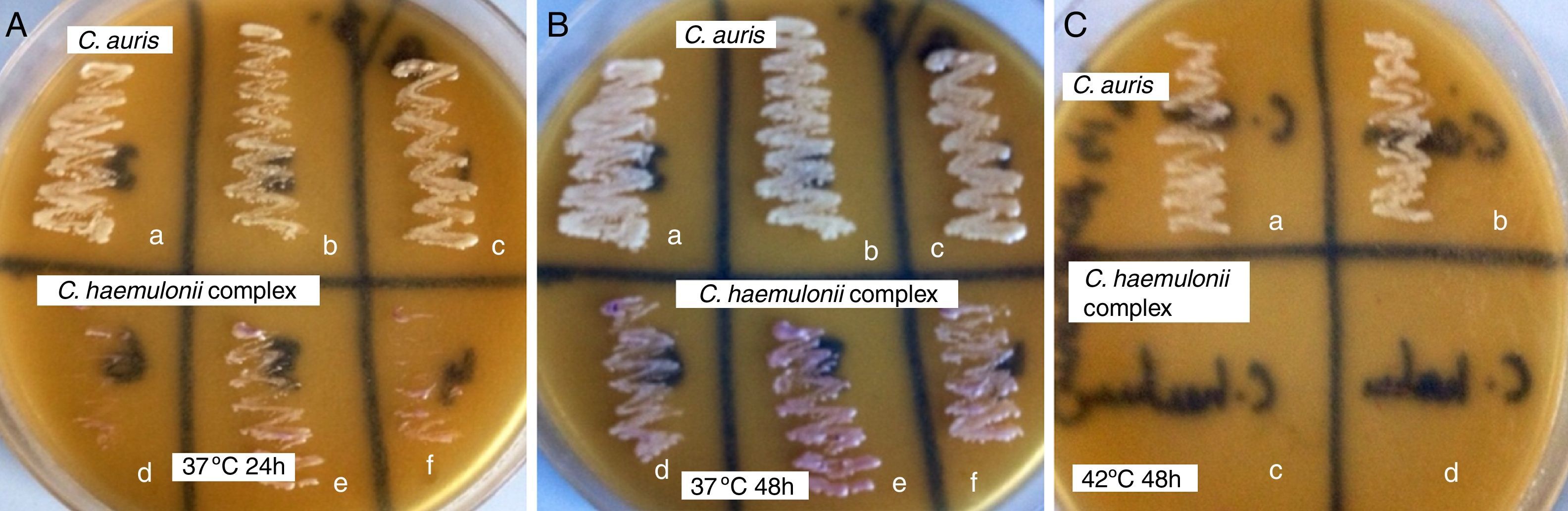
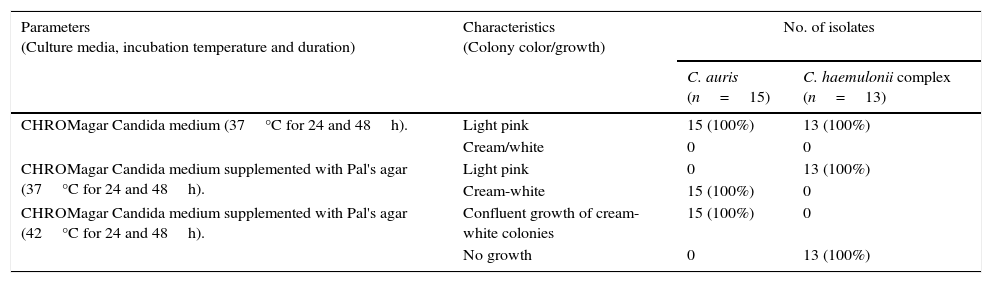
ResultsOn CHROMagar Candida medium supplemented with Pal's agar all C. auris strains showed confluent growth of white to cream colored smooth colonies at 37°C and 42°C after 24 and 48h incubation and did not produce pseudohyphae. The isolates of the C. haemulonii complex, on the contrary, showed poor growth of smooth, light-pink colonies at 24h while at 48h the growth was semiconfluent with the production of pseudohyphae. C. haemulonii complex failed to grow at 42°C.
ConclusionsWe report a rapid and cheap method using CHROMagar Candida medium supplemented with Pal's agar for differentiating C. auris from isolates identified as C. haemulonii by VITEK2.
Candida auris es una especie única debido a su resistencia a múltiples fármacos y a la identificación errónea por sistemas comerciales como Candida haemulonii. Su correcta identificación es importante para evitar un tratamiento inadecuado.
ObjetivosDesarrollar un método de bajo coste para diferenciar C. auris de aislamientos identificados como C. haemulonii por el método VITEK®2.
MétodosQuince aislamientos de C. auris, seis de C. haemulonii, seis aislamientos de Candida duobushaemulonii y un aislamiento de Candida haemulonii var. vulnera se sembraron en medio CHROMagar Candida enriquecido con medio de Pal para una mejor diferenciación.
ResultadosEn el medio CHROMagar Candida enriquecido con medio de Pal, todos los aislamientos de C. auris presentaron un crecimiento confluente con colonias lisas de color blanco a blanco amarillento a 37 y 42°C tras 24 y 48 h de incubación; no hubo producción de seudohifas. Los aislamientos del complejo C. haemulonii, en cambio, mostraron un crecimiento menor. Las colonias eran lisas y de un color rosa claro a las 24 h; a las 48 h el crecimiento fue semiconfluente y se observó producción de seudohifas. No hubo crecimiento de ninguno de los aislamientos a 42°C.
ConclusionesEl medio CHROMagar Candida enriquecido con medio de Pal es rápido y de bajo coste, y resulta efectivo para identificar C. auris entre cepas previamente identificadas como C. haemulonii por el método VITEK®2.
The first reports about Candida auris were exclusively from external ear infections and since then this yeast has been isolated from a wide variety of samples like blood, tissue diabetic foot infections, nails and broncho-alveolar lavage.2,7,8,14 The resistance of C. auris to fluconazole, as well as the reduced susceptibility to voriconazole and itraconazole and its ability for clonal transmission, is worrisome.3,10 Automated identification systems like VITEK2 (bioMérieux, Marcy I’Etoile, France) are known to misidentify C. auris as Candida haemulonii or Candida famata, while API20C AUX (bioMérieux) misidentifies it as Rhodotorula glutinis.2,3,6,7 Reliable identification of these phylogenetically closely related species can be achieved using DNA sequencing and Matrix-Assisted Laser Desorption Ionization-Time Of Flight Mass spectrometry (MALDI-TOF MS) assays, which unfortunately are not available in most clinical microbiology laboratories. Since the treatment strategies for invasive candidiasis is based on the correct species identification, a wrong identification coupled with misleading high minimum inhibitory concentrations (MIC) limits the therapeutic options. Pal's agar, which was developed for the identification of Cryptococcus neoformans, has been previously used in combination with CHROMagar Candida for differentiating between Candida albicans and Candida dubliniensis.11,12 Therefore we evaluated the use of CHROMagar Candida medium supplemented with Pal's agar to differentiate between C. auris and C. haemulonii complex for all isolates identified as C. haemulonii with VITEK2 system.
Fifteen C. auris (CBS 12822, 12883, 12886, 12885, 12876, 12879, 12881, 12880, 12889, 12875, 12874, 12877, 12878, 12887), 6 isolates each of C. haemulonii (VPCI-1158/P/13, 1164/P/13, 1165/P/13, 716/P/14, 1161/P/13, 1163/P/13) and Candida duobushaemulonii (VPCI-713/P14,1160/P/13,1157/P/13470/P/15,1159/P/13,1162/P/13), and one solitary isolate of C. haemulonii var. vulnera (VPCI 715/P/14) from our previous study were used.6 All isolates’ identification were confirmed by sequencing the ITS region of the ribosomal subunit and D1/D2 region of the large ribosomal subunit (LSU) respectively.2,3,6 All our clinical isolates were initially identified as C. haemulonii by VITEK2 system. Pal's agar was prepared by autoclaving the mixture containing agar, creatinine, glucose, KH2PO4 and sunflower seed extract.11 CHROMagar (Becton Dickinson, Sparks, MD) supplemented with Pal's medium was prepared as described previously by mixing equal volumes of both the medium at their normal strength.12 Test isolates were inoculated on the CHROMagar-Pal's medium after being grown on Sabouraud dextrose agar for 48h. The plates were incubated at 37°C and 42°C and colony characteristics were recorded at 24 and 48h by two different observers. Pseudohyphae formation was detected microscopically using lactophenol cotton blue stain.
The colony color, morphology, growth at 37°C and 42°C and pseudohyphae formation when grown in CHROMagar Candida medium and CHROMagar Candida medium supplemented with Pal's agar have been listed in Table 1. On CHROMagar Candida medium supplemented with Pal's agar all C. auris strains showed confluent growth of white to cream colored smooth colonies at 37°C and 42°C after 24 and 48h incubation and did not produce pseudohyphae. C. haemulonii complex, on the contrary, showed poor growth of smooth light pink colonies at 24h while at 48h the growth was semiconfluent (Fig. 1A and B) with production of pseudohyphae. When incubated at 42°C in CHROMagar Candida supplemented with Pal's medium all C. haemulonii complex isolates failed to grow (Fig. 1C).
Colony color and growth characteristics of C. auris and C. haemulonii complex isolates on different culture media at different incubation temperatures.
| Parameters (Culture media, incubation temperature and duration) | Characteristics (Colony color/growth) | No. of isolates | |
|---|---|---|---|
| C. auris (n=15) | C. haemulonii complex (n=13) | ||
| CHROMagar Candida medium (37°C for 24 and 48h). | Light pink | 15 (100%) | 13 (100%) |
| Cream/white | 0 | 0 | |
| CHROMagar Candida medium supplemented with Pal's agar (37°C for 24 and 48h). | Light pink | 0 | 13 (100%) |
| Cream-white | 15 (100%) | 0 | |
| CHROMagar Candida medium supplemented with Pal's agar (42°C for 24 and 48h). | Confluent growth of cream-white colonies | 15 (100%) | 0 |
| No growth | 0 | 13 (100%) | |
Plates of CHROMagar Candida supplemented with Pal's medium showing confluent growth of white colored colonies of C. auris (a, b and c) and poor growth of light pink colored colonies C. haemulonii var. vulnera (d), C. duobushaemulonii (e) and C. haemulonii (f) after incubation at (A) 37°C for 24h, (B) 37°C for 48h and (C) white colored colonies of C. auris (a, b) and no growth of C. duobushaemulonii (c) and C. haemulonii (d) after incubation at 42°C for 48h.
A false impression of high incidence of C. haemulonii infections in India has been created due to studies based on the identification using VITEK2 system.9,13,15 A recent study reported that in addition to misidentifying these isolates, VITEK2 system also reports higher MIC values for amphotericin B, resulting in inappropriate treatment.7C. auris is phylogenetically very closely related to C. haemulonii complex, the latter comprising the genotypically distinguishable species C. haemulonii, C. duobushaemulonii and C. haemulonii var. vulnera.1,13 Differential media like CHROMagar Candida can presumptively identify Candida albicans, Candida tropicalis and Candida krusei. Colony morphology on CHROMagar Candida is not discriminatory as both C. auris and C. haemulonii complex grew well, producing light pink colonies.2,3,6 Many non-C. albicans Candida species like C. krusei, Candida glabrata, Candida kefyr, Candida guilliermondi and Candida parapsilosis also produce light pink colonies on CHROMagar Candida and CHROMagar Candida supplemented with Pal's medium.4,5C. auris grows well at 37°C and 42°C and does not produce pseudohyphae on corn meal agar after 4–8 days incubation at 28°C.6 In contrast C. haemulonii complex produces pseudohyphae and failed to grow at 42°C.6 The differentiation between C. auris and C. haemulonii complex was achieved by observing the colonies color and their ability to grow when incubated at 42°C in CHROMagar Candida supplemented with Pal's medium with 100% sensitivity and specificity. It is a cheap and rapid method to differentiate these two phylogenetically closely related species, as the alternative methods are DNA sequencing and MALDI-TOF MS assays, which are prohibitively expensive, technically challenging and not available in most clinical microbiology laboratories. The main limitation of this study is that preliminary identification needs to be done by an automated identification system like VITEK2 to rule out other non-C. albicans Candida species that can produce pink colonies on CHROMagar Candida medium.
Conflict of interestWe certify that there are no entities with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript.
Source of fundingNone declared.