Candida auris and Candida haemulonii are emerging and multiresistant pathogens. C. auris has produced hospital outbreaks and is misidentified by phenotypic-based methods. The only reliable identification methods are DNA sequencing and MALDI-TOF.
AimsTo develop a classical-PCR method capable of rapidly and accurately identify C. auris and C. haemulonii.
MethodsA multiplex PCR was carried out in one tube that included an internal control and oligonucleotides that specifically hybridize to the ITS2 region of C. auris and C. haemulonii. The usefulness of the new method was verified by testing a collection of 50 strains of 20 different species (previously identified by ITS sequencing). The selection of species was made in order to emulate the C. auris panel used by the CDC to validate diagnostic tools. In addition, other yeast species not included in the aforementioned panel were incorporated based on reported identification errors.
ResultsThe results obtained with the proposed protocol were in total agreement with those obtained by ITS sequencing.
ConclusionsWe present a PCR method able to unequivocally identify C. auris and differentiate it from C. haemulonii. It is inexpensive, fast and it could be a useful tool to reduce the chances of a C. auris outbreak.
Candida auris y Candida haemulonii son patógenos emergentes y multirresistentes. C. auris ha sido responsable de brotes hospitalarios y no se puede identificar por métodos fenotípicos. Los únicos métodos de identificación confiables incluyen la secuenciación y el MALDI-TOF.
ObjetivosDesarrollar un método de PCR clásica capaz de identificar rápidamente C. auris y C. haemulonii.
MétodosSe llevó a cabo una PCR múltiple en un tubo que incluyó un control interno y oligonucleótidos que hibridan específicamente con la región ITS2 de C. auris y C. haemulonii. Para comprobar la utilidad del método se utilizó una colección de 50 aislamientos de 20 especies diferentes (identificadas por secuenciación del ITS). La selección de especies se hizo con el fin de emular el panel de especies que ofrece el CDC para la correcta identificación de C. auris. Además, se incluyeron especies que son confundidas con C. auris y no están incluidas en el citado panel.
ResultadosLos resultados obtenidos con el protocolo propuesto estuvieron en total acuerdo con los obtenidos por la secuenciación del ITS.
ConclusionesEl método que presentamos es capaz de identificar inequívocamente C. auris y diferenciarla de C. haemulonii. Es barato, rápido y podría ser una herramienta útil para reducir la posibilidad de brotes por C. auris.
Candida auris and Candida haemulonii are closely related multiresistant yeast pathogens that are increasingly reported worldwide.1,6–8,10,11,13C. auris has become an important cause of nosocomial outbreaks. The CDC recommends all infected or colonized patients are isolated from the rest of the hospital population.6,7,9,11,14 In order to comply with these recommendations, the correct identification of this species in clinical laboratories is essential. Phenotypic-based commercial identification systems misidentify C. auris. The two unique recommended methods for C. auris accurate identification are rDNA sequencing and MALDI-TOF.5,9 Nevertheless, these two methods have limitations regarding speed, availability, database update and cost. The objective of this work is to propose an inexpensive single-tube classical PCR method able to quickly identify C. auris and differentiate it from C. haemulonii.
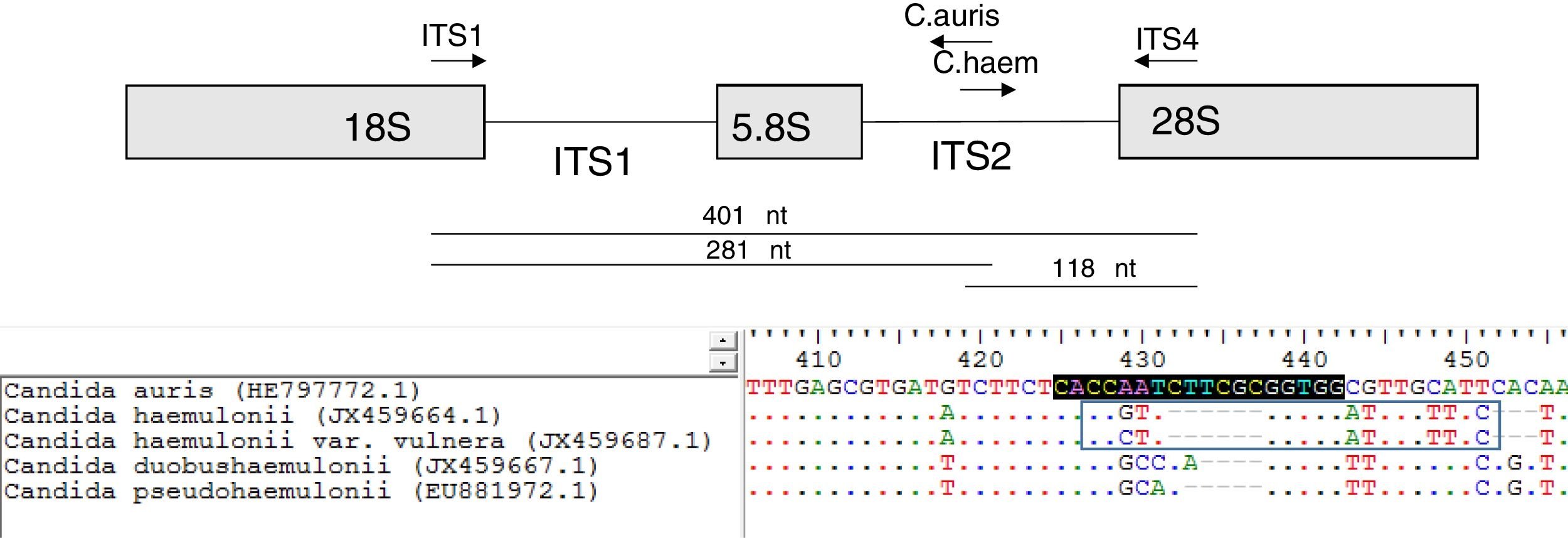
A multiplex PCR tube was assembled with four primers. It contained the ITS universal primers (ITS1 [5′-TCCGTAGGTGAACCTGCGG-3′] and ITS4 [5′-TCCTCCGCTTATTGATATGC-3′]) that were used as internal controls.15 The remaining primers were designed to specifically hybridize the ITS2 region of C. auris and C. haemulonii. These specific primers were named as: C. auris (5′-CCACCGCGAAGATTGGTG-3′; orientation: antisense) and C. haemulonii (5′-CCGTTGGTGGATTTGTTTCT-3′; orientation: sense) (Fig. 1). The primer design was based on the following GeneBank accession numbers: HE797772.1 (C. auris ITS), JX459664.1 (C. haemulonii), JX459687.1 (C. haemulonii var. vulnera), EU881972.1 (Candida pseudohaemulonii) and JX459667.1 (Candida duobushaemulonii). ITS sequences of the species of the C. haemulonii complex were included in the PCR design to ensure the specificity of the primers. Oligonucleotides were purchased from GBT primers (Genbiotech-Argentina). PCR reactions were set in a 25μl final volume following the Pegasus DNA polymerase manufacturer's instructions (PBL-Argentina). Each PCR tube contained 1 unit of Pegasus Taq polymerase, 50ng of yeast DNA, 1× PCR buffer, 250μM dNTPs, 2mM MgCl2, 0.1μM of ITS4, 0.2μM of ITS1 primer and 0.1μM of C. auris and C. haemulonii specific primers. An Applied Biosystems thermocycler was used for DNA amplification using the following program: 2min 95°C (initial step) followed by 25 cycles of 30s at 95°C, 30s at 55°C and 1min at 72°C, and a final step of 10min at 72°C. PCR amplification products were resolved by electrophoresis using a 1.2% agarose gel.
The usefulness of the new method was evaluated by testing a panel of 50 strains of 20 different species specifically assembled for this work. The species were chosen taking into account species that are indistinguishable from C. auris if the currently available methods are used.4,9 Many of these species are included in a CDC panel distributed to be used in the validation of diagnostic tools.3 All the strains of the panel were identified by ITS sequencing15 and were obtained from international collections (e.g. ATCC or CBS) or clinical samples. Our panel included 20 C. auris, 5 C. haemulonii, 1 C. pseudohaemulonii, 1 C. duobushaemulonii, 3 Candida guilliermondii, 2 Candida lusitaniae, 2 Saccharomyces cerevisiae, 1 Rhodotorula glutinis, 1 Candida sake, 1 Candida albicans, 1 Candida glabrata, 2 Candida parapsilosis sensu stricto, 2 Candida krusei, 2 Candida tropicalis, 1 Cryptococcus neoformans, 1 Candida orthopsilosis, 1 Candida nivariensis, 1 Candida dubliniensis, 1 Candida kefyr and 1 Candida famata. Yeast genomic DNAs were obtained using a phenol-based method.12
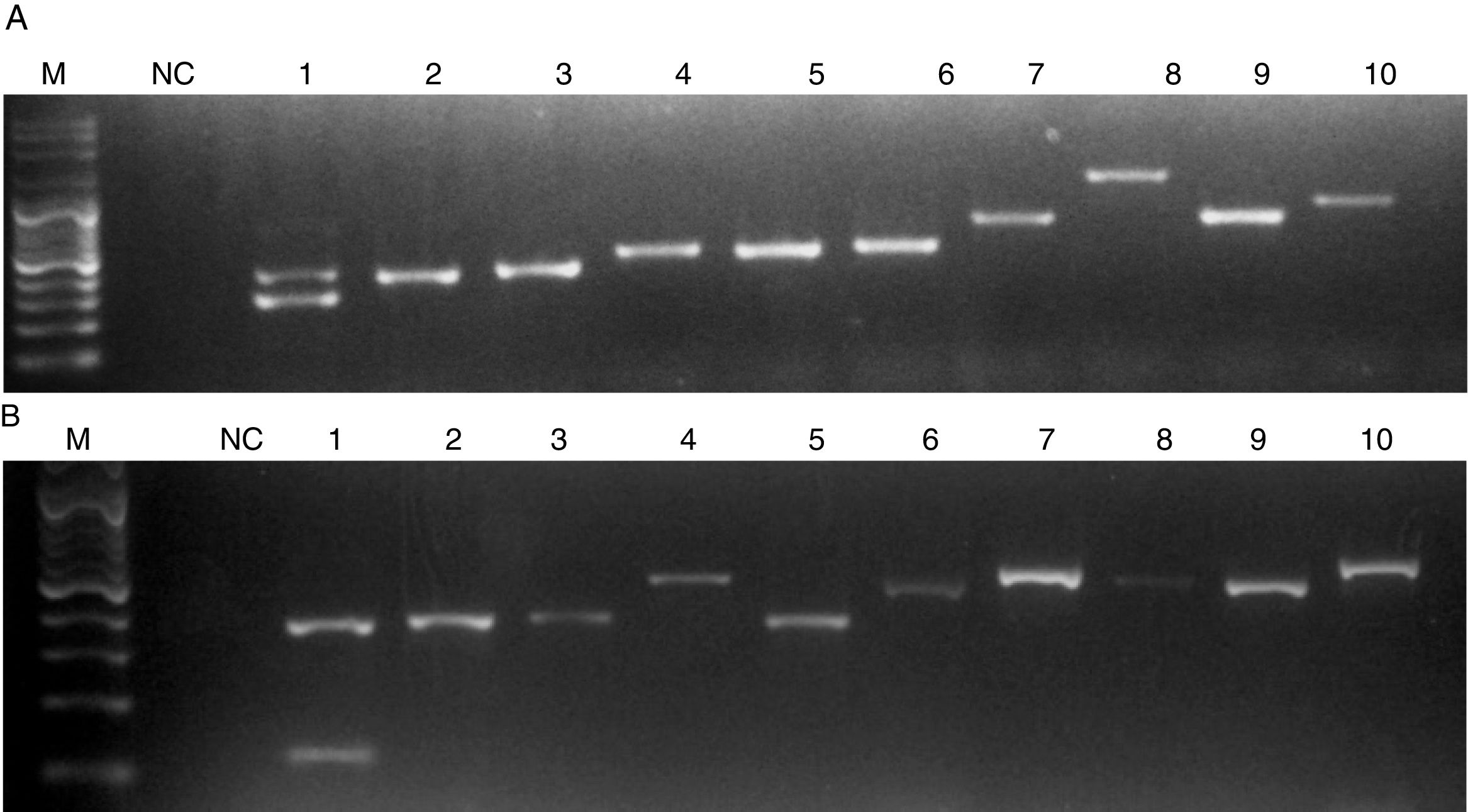
When C. auris or C. haemulonii DNA was used, the PCR amplification yielded two bands: a 401 nt ITS internal control band together with a 281 nt or a 118 nt species specific band, respectively. PCR products of different sizes showing the amplification of the ITS internal controls (using ITS1 and ITS4 primers) were obtained when DNA from other species were used (Fig. 2). In this study, we evaluated 50 strains and the results obtained with the proposed single tube PCR protocol were in total agreement with those obtained by ITS sequencing.
Electrophoresis of the PCRs resolved in a 1.2% agarose gel. M, molecular size marker. NC, negative control. (A) Lanes: 1, C. auris; 2, C. pseudohaemulonii; 3, C. sake; 4, Cryptococcus neoformans; 5, C. krusei; 6, C. dubliniensis; 7, Rhodotorula glutinis; 8, S. cerevisiae; 9, C. famata; 10, C. guilliermondii. (B) Lanes: 1, C. haemulonii; 2, C. pseudohaemulonii; 3, C. duobushaemulonii; 4, C. orthopsilosis; 5, C. lusitaniae; 6, C. albicans; 7, C. neoformans; 8, C. parapsilosis; 9, C. tropicalis; 10, C. nivariensis.
Since the first C. auris isolation, rDNA ITS region sequencing has been used as the unique unequivocal identification tool.5,9,13 Later, the introduction of the MALTI-TOF technology in clinical labs added a new identification option.4 In 2016, the CDC suggested to molecularly characterize or identify by means of MALDI-TOF any clinically resistant yeast and/or any yeast identified as (i) Candida sake or Rhodotorula glutinis (with non-red colony) by using API20C, (ii) Candida catenulata by DB Phoenix and (iii) C. catenulata, C. guilliermondii, C. lusitaniae or Candida famata when using MicroScan.4,9
Herein we present a fast and inexpensive PCR method able to unequivocally identify the multiresistant species C. auris and C. haemulonii. As any other method of molecular identification, a possible drawback is the misidentification due to point mutations in the region where the primers hybridize. However, the ITS regions of all the C. auris and C. haemulonii strains included in this work were sequenced and showed exactly the same nt sequences as the genebank sequences used for the primers design. It must be noted that all the strains used in this work were isolated in different hospitals of Colombia. Geographic clonality was described for C. auris strains.2,9 Thus, these methodology should be tested using C. auris from other world's regions to ensure that all the C. auris share the exact ITS sequence. Other limitation of this PCR technique is that it is not able to differentiate C. haemulonii from C. haemulonii var. vulnera (a variety of C. haemulonii group I).2
The proposed PCR would be a useful tool for supporting the decision to isolate a patient, reducing the possibility of C. auris outbreaks.
Conflict of interestNone declared.
This work was supported in part by the Argentinian Ministerio de Ciencia y Tecnología (MinCyT) [grant number PICT-2013-1571, 2013-2017] to GGE. CD have a postdoctoral fellowship from CONICET.